Abstract
Background
Acute respiratory tract infections (ARTIs) are one of the major causes of morbidity and mortality among young children in developing countries. Information on the incidence of human metapneumovirus (hMPV) and human bocavirus (HBoV) infections in developing countries, especially among rural children, is very limited.
Objectives
This study was conducted to identify whether these viruses were associated with ARTI among children ≤5 years of age in rural and peri-urban populations in South India.
Methods
The study was cross-sectional with prospective sample collection. Oropharyngeal swabs were collected from children ≤5 years of age presenting with ARTI. None of the children in this study were known to have any immunosuppressive conditions. The two viruses, hMPV and HBoV, were identified using semi-nested polymerase chain reaction (PCR) assays and one-step PCR assays, respectively. The lower limits of detection of hMPV and HBoV were 6.69 × 105 plasmid copies and 5.77 × 103 plasmid copies, respectively, per 5 μL PCR reaction input.
Results
The frequency of hMPV infection in children was higher than that of HBoV infection. The different frequencies of hMPV in patients in various age groups with upper and lower respiratory tract infections were compared, and the variance was found to be insignificant. In the 38 children who were hMPV positive, the majority (73.7 %) were from rural communities. The overall hMPV-positive rate was higher in the rural population than in the peri-urban population, but the difference was statistically insignificant. The youngest age at which hMPV-positive status was recorded was 5 months.
Conclusion
This study demonstrated that hMPV was associated with a significant number (i.e. >10 %) of ARTIs in children in South India, whereas a relatively smaller number of HBoV infections was observed.
Similar content being viewed by others
1 Introduction
Acute respiratory tract infections (ARTIs) are a leading cause of illness, especially in pediatric patients. The common respiratory viruses include rhinoviruses, respiratory syncytial virus, influenza virus, adenoviruses, parainfluenza viruses, and coronaviruses [1–3]. However, one third of lower respiratory tract infections (LRTIs) and half of upper respiratory tract infections (URTIs) are left undiagnosed. Since the initial discovery of human metapneumovirus (hMPV) in the Netherlands in 2001, and human bocavirus (HBoV) in Sweden in 2005, these viruses have become recognized as important pathogens responsible for ARTIs in children [4]. These viruses are also important causes of ARTI in the elderly and in immunocompromised patients. hMPV belongs to the Paramyxoviridae family and has a negative-sense single-stranded RNA genome, which includes eight genes coding for nine different proteins [5]. HBoV belongs to the Parvoviridae family and has single-stranded DNA [6]. hMPV and HBoV are now considered important viral pathogens that cause undiagnosed LRTI and URTI in children less than 5 years of age when not tested for [6, 7]. hMPV and HBoV cannot be cultivated reliably in many commonly used cell lines; however, polymerase chain reaction (PCR)-based detection in clinical specimens has been successful. Little is known about the prevalence of hMPV and HBoV among young children from rural communities in South India.
The objective of this study was to analyze the frequency of hMPV and HBoV infection among children ≤5 years of age from rural and peri-urban communities in South India. This article also describes the design and application of a semi-nested reverse transcription PCR assay for detection of hMPV and a one-step PCR assay for detection of HBoV in oropharyngeal swab specimens, and the clinical presentation in different age groups of these young children.
2 Materials and Methods
2.1 Specification of the Target Area
Vellore district lies at a latitude of 12°15′ to 13°15′ North and at a longitude of 78°20′ to 79°50′ East in Tamil Nadu state, India. The district has a large geographical area of 6077 km2. The total area of the city of Vellore is 392.62 km2. Vellore has a tropical wet and dry climate in different months of the year. Summer-like weather arrives in the month of April and persists until the end of June. The humidity ranges from 40 % to 63 % during summer and from 67 % to 86 % during the cooler months. The month of July sees the onset of the monsoon. The temperature ranges from a minimum of 29 °C to a maximum of 42 °C in summer and from a minimum of 12 °C to a maximum of 27 °C in the cooler months.
2.2 Study Population
At the time of the 2011 Census [8], the estimated total population of young children (aged 0–6 years) in Vellore district was 406,705. Of this total, 57.57 % (120,754 boys and 113,390 girls) lived in rural communities and 42.43 % (88,414 boys and 84,147 girls) lived in urban communities. Children aged 0–6 years constituted 10.35 % of the total population in Vellore district. In the Vellore metropolitan area alone, the total population of children aged 0–6 years was 45,049.
Our study population recruitment areas were situated on the outskirts of Vellore city (about 10 km away from the city) and covered primarily peri-urban, rural, and contiguous tribal area populations in and around the southern side of the city.
2.3 Sample Collection
This study was cross-sectional and prospective in nature. Oropharyngeal swabs (n = 300) were collected from as many children ≤5 years of age as possible who sought medical attention for suspected viral acute URTI or LRTI at outpatient clinics at our hospital and in other rural areas of Vellore city. The children were recruited consecutively. None of the children in this study were known to have any immunosuppressive conditions. The sampling sites were the Sri Narayani Hospital and Research Centre (SNH&RC) in Sripuram, Vellore, Vellore Government Medical College and Hospital (VGMCH) in Adukamparai, Vellore, an outreach clinic of SNH&RC in Kaspa, Vellore, a primary health center in Ussoor, Vellore, and an orphanage in Sisubhavan, Vellore.
The sample size was calculated in Epi Info Version 6.04d software (Centers for Disease Control [CDC], Atlanta, GA, USA). Sampling was done during the period of November 2010 to November 2011. Ethical clearance was obtained for the study protocol. Parents were informed as to the study objective, and their written consent was obtained along with completion of a detailed clinical questionnaire before specimen collection.
2.4 Collection of Oropharyngeal Samples
The patient was instructed to open the mouth widely. Using a tongue depressor, the investigator swabbed both the posterior pharynx and the tonsillar areas with sterile Dacron swabs (Puritan Medical Products Company LLC, Guilford, ME, USA), in the clockwise direction, and the swabs were withdrawn (without touching the tongue) as soon as the child gagged, as described in the CDC’s specimen collection guidelines [9]. The swabs were immediately placed in a sterile tube containing 2 mL of Hank’s Balanced Salt Solution (HBSS) [Gibco, Grand Island, NY, USA], mixed well, transported to the laboratory as quickly as possible in ice boxes, and stored at −20 °C until further processing. The PCR assay for HBoV (DNA virus) was done within 2 weeks of sample collection.
2.5 Reverse Transcription PCR Assay for hMPV
RNA extraction was carried out on the same day as the sample collection, using a QIAamp viral RNA mini kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer’s instructions, and stored at −20 °C. A semi-nested reverse transcription PCR assay for hMPV was carried out immediately to avoid RNA decay.
Reverse transcription and subsequent complementary DNA (cDNA) amplification in a semi-nested format was carried out for amplification of a portion of the nucleoprotein (N) gene of hMPV, using a OneStep RT-PCR kit (Qiagen GmbH). The thermal cycling conditions of the external round included 50 °C for 30 min (the reverse transcription step), 95 °C for 15 min, 35 cycles of 95 °C for 1 min, 52 °C for 45 sec, 72 °C for 1 min, and final extension at 72 °C for 7 min. The thermal cycling conditions in the internal round were 95 °C for 15 min, 35 cycles of 95 °C for 1 min, 53 °C for 45 sec, 72 °C for 1 min, and final extension at 72 °C for 7 min. The amplicon size in the internal round was 365 bp.
2.6 One-Step PCR Assay for HBoV
The PCR mix for each reaction contained HotStar Taq polymerase (2U; Qiagen GmbH), deoxyribonucleotide triphosphates (dNTPs) mix (2.5 mM each) [Fermentas Inc., MD, USA], forward and reverse primers (25 pmol) [MetabionInternational AG, Martinsried, Germany], and the template (5 μL). The thermal cycling conditions were 95 °C for 15 min, 35 cycles of 95 °C for 1 min, 54 °C for 45 sec, 72 °C for 1 min, and final denaturation for 7 min at 72 °C. The amplicon size in the one-step PCR was 354 bp. Primers for hMPV and HBoV are shown in Table 1.
2.7 Nested PCR Assay for HBoV
Fifty representative samples (sample nos. 250–300) were tested using a nested PCR format. The internal primers were selected within the external amplification region using Primer 3 software. The amplicon size in the nested PCR was 182 bp.
2.8 Safety Precautions
All infectious specimens were handled in a biosafety level II hood. All precautions were taken for PCR testing, such as flow through, disposable plastic ware, filter blocked tips, and dedicated micropipettes. Micropipettes with aerosol-free filter blocks were used to avoid cross-contamination. Disposable gloves were used and changed between samples. An adequate number of negative controls (‘no-template’ controls) was included as every third sample, along with positive controls in all of the assays. PCR was carried out in a Mastercycler® Personal 5332 thermal cycler (Eppendorf, Hamburg, Germany).
An aliquot of 5 μL of amplicon mixed with 5 μL of 6X Loading Dye (Fermentas Inc., Glen Burnie, MD, USA) was analyzed by gel electrophoresis in 2 % agarose gels (Sigma, St. Louis, MO, USA) prepared in Tris-Borate-EDTA buffer containing 0.5 μg/mL of ethidium bromide (Sigma). A molecular weight marker (100 bp ladder) [Fermentas Inc.] was used in every run. The gels were examined in a gel documentation system (Genei, Bangalore, India) for amplification products.
2.9 Establishment of the Lower Limit of Detection of hMPV and HBoV Species by Plasmid Titration
Plasmids containing the whole virus (HBoV1 nt 1–5299) that was cloned into XhoI/XbaI digested pBluescript SK(+) vector (Stratagene), which resulted in pHBoV1 and the plasmids containing hMPV N gene, were kindly provided by Dr. Eric Simoes. The plasmids were used for examining the lower limit of detection and as positive controls in every PCR run.
The probability of detecting hMPV and HBoV plasmids in a suspension of a known concentration in the presence of defined DNA copy numbers was determined essentially as described previously [10]. The lower limit of detection for PCR was determined using plasmid dilutions and appropriate negative controls (nuclease-free water). A negative control was included after each triplicate of every dilution tested. Amplification shown in the highest dilution (the lowest concentration) in at least two replicates of the triplicates tested at each dilution was taken as the lower limit of detection as plasmid copies/μL. The approximate number of plasmid copies/μL of DNA suspension was thus established. The lower limits of detection of the semi-nested PCR assay for hMPV and the one-step PCR assay for HBoV were 6.69 × 105 plasmid copies and 5.77 × 103 plasmid copies, respectively, per 5 μL PCR reaction input. The lower limit of detection of the HBoV nested PCR was 5.77 plasmid copies per 5 μL PCR reaction input.
Statistical analyses were performed using Epi Info Version 6.03 software.
3 Results
From November 2010 to November 2011, a total of 300 oropharyngeal samples were collected from children who attended the pediatric outpatient clinics at the sampling sites, presented with respiratory symptoms, and met the study inclusion criteria. Table 2 shows the various sampling sites and the numbers of samples collected at each site. Of the 300 children, 172 (57.33 %) were male and 128 (42.67 %) were female.
Eighty-eight patients (29.33 %) were from peri-urban communities (48 male [54.55 %] and 40 female [45.45 %]) and 212 patients (70.67 %) were from rural communities (124 male [58.49 %] and 88 female [41.51 %]). The ages of the patients ranged from 1 to 60 months (mean 29.61 months). Among the samples that were collected, 38 (12.68 %) were positive for hMPV on semi-nested PCR and two (0.67 %) were positive for HBoV on one-step PCR. The two HBoV-positive patients were negative for hMPV. A gel picture showing amplification of hMPV-specific and hBoV-specific genes in patient samples is shown in Fig. 1. Of the 38 hMPV-positive patients, nine had LRTI and 29 had URTI. Of the two HBoV-positive patients, one had URTI and the other had the clinical presentation of LRTI. The frequencies of hMPV in patients indifferent age groups with URTI and LRTI were compared and found to be insignificant. Of the 38 hMPV-positive patients, a high proportion (73.68 %) were from the rural population, while only 26.32 % were from the peri-urban population. When the hMPV-positive rates in rural patients (13.2 %) and peri-urban patients (11.3 %) were compared, the difference was statistically insignificant (p = 0.8). The difference in hMPV-positive status between rural males (12 %) and urban males (16.7 %) was also statistically insignificant (p = 0.6). Thus, the overall hMPV-positive rate did not differ significantly between the rural and peri-urban populations.
Gel picture showing amplification of a human metapneumovirus (hMPV) semi-nested polymerase chain reaction (PCR) and b human bocavirus (hBoV) one-step PCR in patient samples. a Lanes 1 and 4 patient samples showing specific amplification for hMPV at 365 bp; lane 16 positive control; lane 17 molecular weight marker (100 bp ladder). b Lane 1 patient sample showing specific amplification for HBoV; lane 10 positive control; lane 11 molecular weight marker (100 bp ladder)
The majority of the children had cough (94.3 %) and coryza (88.6 %). The four symptoms most commonly found in our study were dry cough, cough with secretions, coryza, and rhinitis, but overall these symptoms were not significantly associated with hMPV-positive status (Table 3). The frequencies of individual symptoms in children in different age groups were analyzed and are shown in Table 4. Symptoms that were significantly more common in hMPV-positive children were coryza in children aged ≥24 months, rhinitis in children aged ≥12 months, vomiting in children aged 24–35 months, and cough with secretions in children aged ≥36 months. The youngest ages at which hMPV-positive and HBoV-positive status was recorded were 5 months and 11 months, respectively.
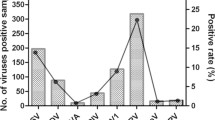
The frequencies of hMPV-positive and hMPV-negative ARTIs in different months of the year were compared. hMPV-positive ARTIs were more common during the cooler and wetter months (July to January) [Fig. 2]. hMPV-positive infections were more common than HBoV-positive infections. All PCR amplicons that were positive on PCR were sequenced and subjected to Basic Local Alignment Search Tool (BLAST) analysis. Analysis of the clustal-aligned sequences confirmed the presence of the respective viruses. The N gene of hMPV was used to distinguish genotypes, and a phylogenetic tree including comparison with global strains is being reported separately. When a subset of 50 samples (sample nos. 250–300) were tested for hBOV, using the nested PCR format, one sample was additionally shown to be positive for HBoV. The PCR amplicon was sequenced, subjected to a BLAST search, and found to be 100 % similar to the existing HBoV sequence in the GenBank repository.
4 Discussion
Respiratory tract infections caused by hMPV and HBoV are a major cause of concern for the health of infants and young children [11]. The present article presents data on the frequencies of hMPV and HBoV infections in a prospective cross-sectional study of children ≤5 years of age. In this cross-sectional study, we collected oropharyngeal samples for detection of these two viruses. The age and domiciliary status (peri-urban or rural) of each patient was recorded in order to assess the distribution of these viruses in different age groups and different domiciles.
Previous studies in different countries have focused on the involvement of both hMPV and HBoV in respiratory tract infections in children [12–15] and in immunocompromised adults [16].
Côté et al. [17] evaluated detection of hMPV using real-time PCR assays targeting different regions (the N, M, L, F, and P genes), and found that the N and L genes that code for internal viral proteins were particularly suitable for hMPV detection with high specificity and sensitivity.
The viral capsid proteins (VP1/VP2) and non-structural protein (NS-1 and NP-1) coding genes are generally used as target genes for detection of HBoV. Schildgen et al. [11], however, found VP1/VP2 to be a highly variable region and the NS-1 and NP-1 genes to be more conserved. We therefore used the N gene of hMPV and the NP-1 gene of HBoV for both amplification and sequence confirmation in our study.
Though direct fluorescent antibody testing and rapid cell culture can be performed for hMPV, they are not done routinely [18]. Molecular assays such as PCR have been widely used for detection of these viruses in respiratory samples. Respiratory virus detection is highly dependent on the type of sample collected, in addition to the time of collection, the age of the patient, and the storage of samples prior to testing. Samples such as nasopharyngeal aspirates, nasopharyngeal washes, and nasopharyngeal swabs have been reported to be the best samples [18].
Maggi et al. [19] demonstrated hMPV in nasal swabs and in blood plasma by PCR, with a highest positive rate of 43 %. Nasopharyngeal swabs have a slightly higher rate of sensitivity than throat swabs (90 %) in isolation of hMPV [20]. In our study, collection of nasopharyngeal samples was difficult in terms of both non-compliance by parents and non-cooperation by children. Thus, instead of simply collecting throat swabs, we collected oropharyngeal swabs. In our opinion, these swabs should be of adequate sensitivity for virus detection. This approach has been found to be satisfactory for detection of respiratory viruses [21, 22]. Kim et al. [23] recently assessed the relative performance of nasopharyngeal and/or oropharyngeal swab specimens from pediatric and adult patients. The two methods did not differ significantly for detection of human metapneumovirus and other viruses such as influenza A (H3N2) virus, parainfluenza virus 1, or respiratory syncytial virus.
In a study using nested PCR for both hMPV and HBoV, Guido et al. [24] identified hMPV and HBoV with positive rates of 18.9 % and 45.7 %, respectively, in oropharyngeal swabs collected from children.
In our study, the frequencies of hMPV and HBoV were 12.7 % and 0.67 %, respectively. The frequency of hMPV in our study was consistent with those reported in other parts of the world. A semi-nested PCR approach was used for detection of hMPV, and a one-step format was used for detection of HBoV. The low positivity rate of HBoV could be due to the non-nested type of PCR that was undertaken. However, since we identified only one more hBOV-positive patient by use of nested PCR in the 50 subset samples, we believe that the frequency of HBoV may have been low in our study population.
We found that the number of hMPV-positive samples was higher in patients tested at VGMCH than in those tested at other study sites. The number of samples collected from VGMCH was higher than at the other sites, and we believe that such government hospitals are generally preferred by patients at acute stages of infection, particularly patients from rural communities. Moreover, the majority of the patients were from low socioeconomic status groups. The difference in the frequencies of hMPV-positive samples collected at VGMCH and SNH&RC was, however, statistically insignificant (χ2 = 1.76; p = 0.18).
The risk factors for viral respiratory infection are prematurity, bottle feeding, overcrowding, domestic smoke pollution due to the use of woodstoves, and lack of sinks in the household [25–27].These risk factors are very commonly seen in Indian rural areas. In our study, more patients were from rural areas than from peri-urban areas.
Widespread antimicrobial use in the treatment of hospitalized patients creates an ideal environment for development of antimicrobial resistance [28]. This could be limited by use of PCR assays that establish the cause of ARTI in children, including hMPV and HBoV. Rapid identification of viral infections can, furthermore, help to control secondary bacterial community-acquired pneumonia and nosocomial transmission [29].
hMPV has been reported to be an important etiologic agent in LRTI, i.e. in bronchiolitis, exacerbations of asthma/wheezing, pneumonitis in transplant recipients and, rarely, pneumonia [30–32]. Manoha et al. [33] found no difference in the prevalence of bronchiolitis in children with hMPV, respiratory syncytial virus, and rhinovirus. Though hMPV has also been reported to be associated with URTI [34, 35], such reports have either presented a very low prevalence rate (i.e. 5 %) or indicated that the virus was more commonly associated with LRTI than with URTI. hMPV infection associated with URTI was reported to be restricted to reinfection in adults [36, 37]. HBoV has been largely reported to be an important causative agent primarily of LRTI in children [38–40]. In our study, we included both URTI and LRTI as inclusion criteria, and we compared HBoV status in different age groups, but the difference was statistically insignificant in all age groups. The HBoV detection rate was too low for a meaningful comparison between groups to be made.
The hMPV-positive rate has been reported to be higher at the start of the warmest part of the year [34, 41, 42], and other studies have shown the virus during the coolest part of the year [43, 44]. In our study, we found a higher positivity rate of hMPV during both the cooler and wetter months of the year. The varying pattern of hMPV circulation may be due to climatic and demographic differences.
Apart from pneumonia and bronchiolitis, symptoms such as wheezing, asthma, difficulty breathing, and sore throat have been reported to be more common in patients with hMPV infections [45, 46]. In our study, we found that dry cough, cough with secretions, coryza, and rhinitis were more common in hMPV-positive patients than in hMPV-negative patients. Overall, however, the difference was not statistically significant for any symptom. However, when the frequencies of each symptom were compared between hMPV-positive and hMPV-negative patients in each age group, coryza was significantly more common in hMPV-positive children aged ≥2 years and rhinitis was significantly more common in hMPV-positive children aged ≥1 year. Vomiting and cough with secretions were significantly more common in hMPV-positive patients aged 2–3 years and ≥3 years, respectively.
Hajjar et al. [47] found that the incidence of hMPV was 8.3 % among Saudi children. The infection was prominent during the autumn and winter. Zhu et al. [48] found that the prevalence of hMPV infection was 4 % among children tested in Beijing in a surveillance study during a 4-year period. The peak of hMPV activity mostly occurred in late spring, and the majority of patients had LRTI. Most hMPV-positive children were under 5 years of age.
Bharaj et al. [1] found hMPV in 3.6 % of children in a study using multiplex PCR for detection of viruses in North India. In a recent study from the same hospital, by Banerjee et al. [49], the seroprevalence of hMPV was higher in adults than in children aged <5 years. However, the sample size of each age group in the latter study was too small to allow meaningful comparisons. Moreover, it is possible that antibody detection could be less sensitive than RNA detection in respiratory infections. Banerjee et al. [50] reported a 12 % frequency of hMPV infections in North India, and Agrawal et al. [51] reported a 5 % frequency in eastern India.
To date, there are only limited data available on hMPV among children in India, and this article is the first to report the frequency of hMPV and HBoV in South India, particularly in rural and peri-urban children.
In our study, the youngest age at which hMPV-positive status was recorded was 5 months. Four patients had wheezing, but none of the patients whose samples tested positive for hMPV had asthma, rashes, or diarrhea. Rawlinson et al. [52] found that hMPV was less frequent in children with asthma. Fujitsuka et al. [53] also found that hMPV was very rare in Japanese children with acute wheezing illness. The link between hMPV and the induction of wheezing and exacerbation of asthma is still not clear.
In our study, the detection limit of the semi-nested PCR targeting the N gene was 6.69 × 105 plasmid copies per reaction. Ali et al. [54] reported that the MultiCode-PLx multiplex assay was more sensitive than an individual real-time reverse-transcription PCR assay for detection of 11 common respiratory viruses, including hMPV. The MultiCode-PLx multiplex assay, though very rapid and sensitive, is highly expensive and is not commonly available in developing countries such as India.
Our study had limitations. The patients shown to be positive for hMPV or HBoV were not tested for co-infection with other viruses, such as respiratory syncytial virus. Furthermore, no (endogenous) ‘housekeeping gene’ was amplified to rule-out false negative PCR findings.
5 Conclusion
This study demonstrated that hMPV is associated with a considerable number of ARTI cases among young children in South India, whereas a relatively smaller number of HBoV infections were observed in these children. Our data underline the role of hMPV in the causation of ARTI in South Indian children aged ≤5 years from rural and peri-urban populations. Further studies are needed for better understanding of the epidemiology of these viruses in our population in large field studies and hospital-based studies including screening for a number of respiratory viruses.
References
Bharaj P, Sullender WM, Kabra SK, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89.
Mahony J, Chong S, Merante F, et al. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid micro bead-based assay. J Clin Microbiol. 2007;45(9):2965–70.
Stempel HE, Martin ET, Kuypers J, et al. Multiple viral respiratory pathogens in children with bronchiolitis. Acta Paediatr. 2009;98(1):123–6.
Kleines M, Scheithauer S, Rackowitz A, et al. High prevalence of human bocavirus detected in young children with severe acute lower respiratory tract disease by use of a standard PCR protocol and a novel real-time PCR protocol. J Clin Microbiol. 2007;45(3):1032–4.
Kahn JS. Epidemiology of human metapneumovirus. Clin Micro Rev. 2006;19(3):546–57.
Allander T, Tammi MT, Eriksson M, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–6.
Williams JV, Harris PA, Tollefson SJ, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350(5):443–50.
Population census India. http://www.census2011.co.in. Accessed 2012 Oct 17.
Centers for Disease Control [CDC]. Specimen collection guidelines. http://emergency.cdc.gov/urdo/pdf/SpecCollectionGuidelines.pdf. Accessed 2012 Oct 17.
Malorny B, Hoorfar J, Bunge C, et al. Multicenter validation of the accuracy of Salmonella PCR: towards an international standard. Appl Environ Microbiol. 2003;69(1):290–6.
Schildgen O, Müller A, Allander T, et al. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin Microbiol Rev. 2008;21(2):291–304.
Smuts H, Workman L, Zar HJ. Role of human metapneumovirus, human coronavirus NL63 and human bocavirus in infants and young children with acute wheezing. J Med Virol. 2008;80(5):906–12.
Milder E, Arnold JC. Human metapneumovirus and human bocavirus in children. Pediatr Res. 2009;65(5 Pt 2):78R–83R.
Pilger DA, Cantarelli VV, Amantea SL, et al. Detection of human bocavirus and human metapneumovirus by real-time PCR from patients with respiratory symptoms in Southern Brazil. Mem Inst Oswaldo Cruz. 2011;106(1):56–60.
Zappa A, Canuti M, Frati E, et al. Co-circulation of genetically distinct human metapneumovirus and human bocavirus strains in young children with respiratory tract infections in Italy. J Med Virol. 2011;83(1):156–64.
Müller A, Klinkenberg D, Vehreschild J, et al. Low prevalence of human metapneumovirus and human bocavirus in adult immunocompromised high risk patients suspected to suffer from Pneumocystis pneumonia. J Infect. 2009;58(3):227–31.
Côté S, Abed Y, Boivin G. Comparative evaluation of real-time PCR assays for detection of the human metapneumovirus. J Clin Microbiol. 2003;41(8):3631–5.
Ginocchio CC, McAdam AJ. Current best practices for respiratory virus testing. J Clin Microbiol. 2011;49(9):44–8.
Maggi F, Pifferi M, Vatteroni M, et al. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J Clin Microbiol. 2003;41(7):2987–91.
Lambert SB, Whiley DM, O’Neill NT, et al. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using teal-time polymerase chain reaction. Pediatrics. 2008;122(3):615–20.
Lieberman D, Lieberman D, Shimoni A, et al. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling. J Clin Microbiol. 2009;47(11):3439–43.
Lieberman D, Lieberman D, Shimoni A, et al. Pooled nasopharyngeal and oropharyngeal samples for the identification of respiratory viruses in adults. Eur J Clin Microbiol Infect Dis. 2010;29:733–5.
Kim C, Ahmed JA, Eidex RB, et al. Comparison of nasopharyngeal and oropharyngeal swabs for the diagnosis of eight respiratory viruses by real-time reverse transcription-PCR assays. Plos One. 2011;6(6):1–6.
Guido M, Quattrocchi M, Campa A, et al. Human metapneumovirus and human bocavirus associated with respiratory infection in Apulian population. Virology. 2011;417(1):64–70.
Bulkow LR, Singleton RJ, DeByle C, et al. Risk factors for hospitalization with lower respiratory tract infections in children in rural Alaska. Pediatrics. 2012;129(5):1220–7.
Chauhan AJ, Johnston SL. Air pollution and infection in respiratory illness. Br Med Bull. 2003;68:95–112.
Pandey MR, Neupane RP, Gautam A, et al. Domestic smoke pollution and acute respiratory infections in a rural community of the hill region of Nepal. Environ Int. 1989;15:337–40.
Shiley KT, Lautenbach E, Lee I. The use of antimicrobial agents after diagnosis of viral respiratory tract infections in hospitalized adults: antibiotics or anxiolytics? Infect Control Hosp Epidemiol. 2010;31(11):1177–83.
Berger A, Obwegeser E, Aberle SW, et al. Nosocomial transmission of respiratory syncytial virus in neonatal intensive care and intermediate care units: a prospective epidemiologic study. Pediatr Infect Dis J. 2010;29(7):669–70.
Chen X, Zhang ZY, Zhao Y, et al. Acute lower respiratory tract infections by human metapneumovirus in children in Southwest China: a 2-year study. Pediatr Pulmonol. 2010;45(8):824–31.
Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis. 2011;52(4):S284–9.
Anderson EJ, Simões EAF, Buttery JP, et al. Prevalence and characteristics of human metapneumovirus infection among hospitalized children at high risk for severe lower respiratory tract infection. J Ped Infect Dis. 2012;1:212–22.
Manoha C, Espinosa S, Aho SL, et al. Epidemiological and clinical features of hMPV, RSV and RVs infections in young children. J Clin Virol. 2007;38(3):221–6.
Williams JV, Wang CK, Yang C, et al. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J Infect Dis. 2006;193:387–95.
García-García ML, Calvo C, Martín F, et al. Human metapneumovirus infections in hospitalised infants in Spain. Arch Dis Child. 2006;91(4):290–5.
Boivin G, De Serres G, Hamelin ME, et al. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin Infect Dis. 2007;44(9):1152–8.
Pancer K, Ciacka A, Gut W, et al. Detection of hMPV antigen by EIA in clinical specimens. Przegl Epidemiol. 2011;65(3):415–9.
Ma X, Endo R, Ishiguro N, et al. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44:1132–4.
Zhang LL, Tang LY, Xie ZD, et al. Human bocavirus in children suffering from acute lower respiratory tract infection in Beijing Children’s Hospital. Chin Med J (Engl). 2008;121(17):1607–10.
Ghietto LM, Camara A, Camara J, et al. High frequency of human bocavirus 1 DNA in infants and adults with lower acute respiratory infection. J Med Microbiol. 2012;61(Pt 4):548–51.
Talavera GA, Mézquita ND. Human metapneumovirus in children with influenza-like illness in Yucatan, Mexico. Am J Trop Med Hyg. 2007;76(1):182–3.
Aberle JH, Aberle SW, Redlberger-Fritz M, et al. Human metapneumovirus subgroup changes and seasonality during epidemics. Pediatr Infect Dis J. 2010;29(11):1016–8.
McAdam AJ, Hasenbein ME, Feldman HA, et al. Human metapneumovirus in children tested at a tertiary-care hospital. J Infect Dis. 2004;190(1):20–6.
Regev L, Hindiyeh M, Shulman LM, et al. Characterization of human metapneumovirus infections in Israel. J Clin Microbiol. 2006;44(4):1484–9.
Williams JV, Edwards KM, Weinberg GA, et al. Population-based incidence of human metapneumovirus infection among hospitalized children. J Infect Dis. 2010;201(12):1890–8.
Schildgen V, van den Hoogen B, Fouchier R, et al. Metapneumovirus: lessons learned over the first decade. Clin Microbiol Rev. 2011;24(4):734–54.
Hajjar SA, Thawadi SA, Muhsen SA, et al. Human metapneumovirus and human coronavirus infection and pathogenicity in Saudi children hospitalized with acute respiratory illness. Ann Saudi Med. 2011;31(5):523–7.
Zhu R, Qian Y, Zhao L, et al. Characterization of human metapneumovirus from pediatric patients with acute respiratory infections in a 4-year period in Beijing, China. Chin Med J (Engl). 2011;124(11):1623–8.
Banerjee S, Sullender WM, Ahuja RK, et al. Seroepidemiological study of human metapneumovirus in New Delhi, India. Indian J Med Microbiol. 2011;29(4):363–7.
Banerjee S, Bharaj P, Sullender W, et al. Human metapneumovirus infections among children with acute respiratory infections seen in a large referral hospital in India. J Clin Virol. 2007;38(1):70–2.
Agrawal AS, Roy T, Ghosh S, et al. Genetic variability of attachment (G) and fusion (F) protein genes of human metapneumovirus strains circulating during 2006–2009 in Kolkata, Eastern India. Virol J. 2011;8:67.
Rawlinson WD, Waliuzzaman Z, Carter IW, et al. Asthma exacerbations in children associated with rhinovirus but not human metapneumovirus infection. J Infect Dis. 2003;187(8):1314–8.
Fujitsuka A, Tsukagoshi H, Arakawa M, et al. A molecular epidemiological study of respiratory viruses detected in Japanese children with acute wheezing illness. BMC Infect Dis. 2011;11:168.
Ali SA, Gern JE, Hartert TV, et al. Real-world comparison of two molecular methods for detection of respiratory viruses. Virol J. 2011;8:332.
Acknowledgments
This research work was funded by Indian Council of Medical Research (ICMR) grant no. 5/8/7/15/2009/ECD-I. The authors have no conflicts of interest that are directly relevant to the content of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Narayanan, H., Sankar, S., Simoes, E.A.F. et al. Molecular Detection of Human Metapneumovirus and Human Bocavirus on Oropharyngeal Swabs Collected from Young Children with Acute Respiratory Tract Infections from Rural and Peri-Urban Communities in South India. Mol Diagn Ther 17, 107–115 (2013). https://doi.org/10.1007/s40291-013-0030-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-013-0030-y