Antibiotic Resistance in Black Sea Microbial Communities
- 1Department of Biology and Ecology, State Institution National Antarctic Scientific Center, Kyiv, Ukraine
- 2Faculty of Plant Protection, Biotechnology and Ecology, National University of Life and Environmental Sciences of Ukraine, Kyiv, Ukraine
- 3Environmental Institute, Kos, Slovakia
- 4Laboratory of Analytical Chemistry, Department of Chemistry, National and Kapodistrian University of Athens, Athens, Greece
Background: Antibiotic resistance genes (ARGs) are considered as pollutants and are found in natural and anthropogenically impacted environments. Distribution of ARGs in marine environment poses a threat to human health turning the water body into a pool for the ARGs’ transmission.
Objectives: A large-scale study of antibiotic resistance in microbial communities has been performed in the Black Sea, both in the coastal and offshore regions.
Methods: The quantitative distribution of the genes responsible for the inactivation of the beta-lactam (blaCMY, blaSHV), vancomycin (vanA, vanB), macrolides (ermB) and colistin (mcr-1) was assessed with real-time quantitative PCR. Concentrations of the antibiotics belonging to the classes of beta-lactam/cephalosporin/carbapenem, macrolides and glycopeptides were determined by LC-ESI-QTOF-MS.
Results: The present study revealed the distribution of antibiotic resistance genes targeting the response to all antibiotics included in our analysis at various locations across the Black Sea. According to the ARGs copy number normalized to the 16S rRNA, vanB (2 × 10−1 ± 1 × 10−1) and blaSHV (4 × 10−2 ± 1 × 10−2) were the most numerous genes, followed by blaCMY (1 × 10−2 ± 3 × 10−3) and mcr-1 (3 × 10−2 ± 2 × 10−2). The less abundant gene was ermB (1 × 10−3 ± 5 × 10−4) and vanA (1 × 10−5 ± 5 × 10−4). The mcr-1, blaCMY and blaSHV had moderate positive correlation with markers of ruminant faecal pollution. The concentration of antibiotics in seawater was below the detection limit. The abundance of all ARGs included in the study was significantly higher (p-value<0.05) within the northwest coastal area when compared to the offshore stations. The results clearly indicate an alarming antibiotic resistance problem in the region and call for a regular monitoring of ARGs abundance in the Black Sea and its major freshwater tributaries.
Introduction
The Black Sea is a large semi-closed European sea with an area of 436,400 km2. It is virtually isolated from other seas and ultimately drains into the Mediterranean Sea via the narrow Bosphorus channel. The drainage basin of the Black Sea is ca. 2,000,000 km2 and covers regions with intensive agriculture and industrial activities. Riverine input, especially of the major rivers Danube, Dnieper and Dniester, is a considerable source of the Black Sea pollution (Shimkus and Trimonis, 1974; Alygizakis et al., 2019; Diamanti et al., 2020). Ports and coastal cities contribute to the pollution as well; persistent organic pollutants, metals and pharmaceuticals were frequently found in the Black Sea water, sediment and biota compartments (Bakan and Ariman, 2004; Ozkoc et al., 2007; Stoichev et al., 2007). Considering the overall pollution pattern, contamination of the Black Sea with antibiotic resistance genes (ARG) can be expected (Kümmerer, 2009; Kraemer et al., 2019; Zheng et al., 2021).
Antibiotic resistance is one of the biggest threats to global health, emerging as one of the major causes of death in the coming decades (https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance). ARGs have burst outside the clinic environment long ago (Kraemer et al., 2019), and numerous studies report distribution of ARGs in different water environments (Tang et al., 2015; Szekeres et al., 2018; Griffin et al., 2019) etc. Dissemination of ARGs in the natural environment implies a direct hazard to human health due to the possibility of ARGs and antibiotic resistance bacteria (ARBs) intake with food or by direct contact (Amarasiri et al., 2020; Zheng et al., 2021).
The Black Sea region has already been in focus in terms of the antibiotic resistance., however, most of the studies imply antibiotic resistant testing or ARG profiling of bacteria isolated from fish farms, marine biota, seawater and freshwater from the Danube river (Kayis et al., 2009; Kittinger et al., 2016a; Kittinger et al., 2016b; Türe and Alp, 2016; Terzi and Isler, 2019; Kayis et al., 2021). ARGs distribution in the Black Sea water was studied by Sabatino et al. (2020) in the eastern and western part of the Black Sea. blaCTXM, sul2, and tetA were the most abundant ARGs, being present in at least 43% of the samples, while ermB and qnrS were not detected. A wide-scope chemical screening together with analysis of ARGs was carried out in the effluents of wastewater treatment plants (WWTPs) in the Danube river basin (Alygizakis et al., 2019). According to the study, aph (III)a, blaOXA, ermB, ermF, sul1 and tetM had widespread occurrence, while blaSHV, mecA, qnrS, tetB and vanA were detected sporadically. In general, there is a lack of data on the ARGs distribution in the Black Sea environment.
Antibiotics are considered as important driving factors for ARG proliferation, as they pose selective pressure on microbes and enrich ARGs in estuarine and coastal environments (Zheng et al., 2021). Moreover, antibiotics may encourage the horizontal gene transfer (HGT), which facilitates the dissemination of ARGs (Szczepanowski et al., 2009). Faecal pollution is another factor of ARGs distribution (Khan et al., 2019). According to the publicly available metagenomic data, the presence of ARGs can simply result from faecal contamination with resistant bacteria (Karkman et al., 2019). Poorly treated or untreated sewage discharge containing faecal bacteria may fuel the pool of ARGs in anthropogenically impacted environments, especially in low-income countries.
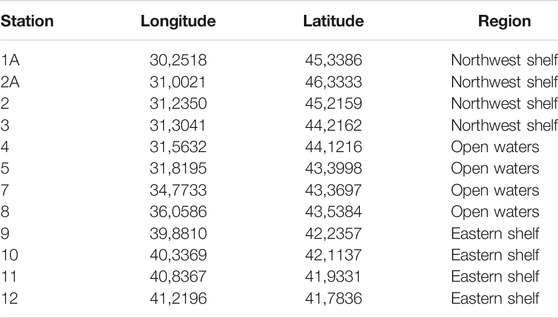
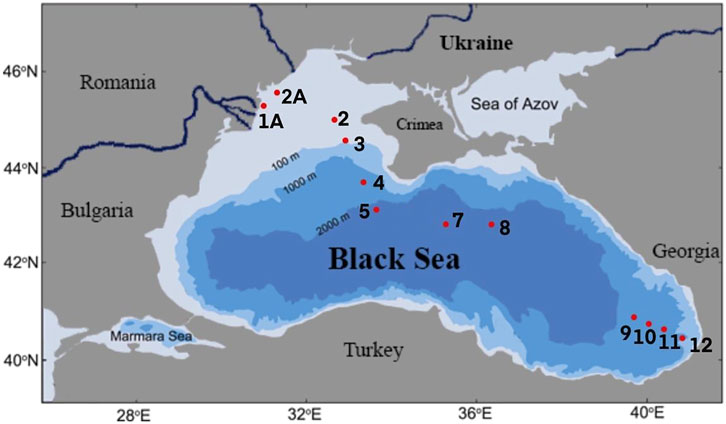
To facilitate an overview of ARGs distribution in the Black Sea environment, 12 water samples were collected in the eastern, central and northwestern parts of the Black Sea in the course of the EU/UNDP EMBLAS-Plus project (https://emblasproject.org/). The research aimed at studying the spatial distribution of ARGs and associated factors contributing to their persistence in the Black Sea. The objectives of this study were to estimate whether: 1) ARGs targeting the antibiotics of wide usage and last-resort antibiotics are present in the Black Sea; 2) there is a difference in ARGs number in the open water and anthropogenically impacted shelf zone of the Black Sea; 3) the number of ARGs is associated with concentration of antibiotics in the water or with the markers of the faecal contamination.
Materials and Methods
Sampling
The samples were collected during the Joint Black Sea Survey on the RV “Mare Nigrum'' carried out within the EMBLAS-Plus project in July and August 2019. Altogether 12 sampling stations were investigated on a transect from Constanta (Romania) to Odessa (Ukraine) and Batumi (Georgia) (Table 1; Figure 1). 2 L of seawater were taken from the surface and passed through the Isopore PC 0.22 µm filters (Millipore, United States) using Microsart e. jet vacuum pump (Sartorius Stedim, Germany). Filters with bacterial biomass on them were immediately frozen at −180°C after filtration.
DNA Extraction
DNA was extracted from seawater using DNAeasy PowerSoil® Kit (Qiagen, Germany) according to the manufacturer’s instructions with the adaptations to the material used. Filters were cut with sterile scissors prior to the lysis step. The DNA quantity and quality were estimated using NanoDrop Spectrophotometer 2000 (Thermo Fisher Scientific, United States). All samples had sufficient DNA concentration (with most above 30 ng/μL) and A260/280 ratio ∼1.8.
Quantitative Real-Time PCR
ARGs that encode resistance to beta-lactams/cephalosporins (blaCMY, blaSHV), vancomycin (vanA and vanB), macrolide-lincosamide (ermB) and colistin (mcr-1) were enumerated by RT qPCR. Primers CMY_fwd/CMY_rev, SHV_fwd/SHV_rev (Roschanski et al., 2014), vanAF/vanAR, vanBF/vanBR (Mirzaei et al., 2015), erm (B)-91f/erm (B)-454r and mcr1FP/mcr1RP (Hembach et al., 2017) were used correspondingly (Supplementary Table S1).
The list of targeted genes was based on the antibiotic resistance prevalence data from the European Resistance Surveillance Network (EARS-NET, 2019) and Joint Danube Survey 3 reports (Zarfel et al., 2015).
Gut host-specific bacteria of the Bacteroidetes phylum were used as markers of faecal pollution. To quantify human Bacteroidetes primers HF183 and BacR287 were used (Epa and of Science, 2019). BacR/BacF (Reischer et al., 2006) and Pig-2-Bac41F/Pig-2-Bac163Rm (Mieszkin et al., 2009) primers were used to quantify ruminant and pig Bacteroidetes respectively. Primers S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 were used to estimate the quantity of the 16S rRNA gene (Klindworth et al., 2013). Primer sequences and annealing temperatures are presented in Supplementary Table S1.
RT qPCR was set using the QuantiFast SYBR Green PCR Kit (Qiagen, Germany) according to the standard manufacturer’s procedures. The standards were created for each of the targeted genes using the DNA from the samples. Each 25 μL PCR reaction contained the following components: 2x QuantiFast SYBR Green PCR Master Mix—12.5 μL, Primer Reverse −2.5 μL, Primer Forward −2.5 μL, template DNA −1 μL, RNase-free water −6.5 μL. The initial concentration of DNA in each reaction was 5 ng/μL. The thermal conditions were different for all primers and are presented in Supplementary Table S1. Negative controls contained no DNA template.
The standards for each gene analyzed were purified with QIAquick PCR Purification Kit (Qiagen, Germany) and 10-fold serially diluted ranging from 1.0 × 103 to 1.0 × 107 to be used for standard curve generation in quantitative PCR. The threshold value (Ct) was used to determine the copy numbers of targeted genes in the environmental subsamples based on the standard curves.
Melting curve analysis was performed at the end of the amplification cycles in order to assess primer specificity and to ensure proper amplification of all target fragments. The PCR reactions were performed on the Qiagen Rotor-Gene Q (Qiagen, Germany). All reactions were performed in triplicate.
Concentrations of Antibiotics
Concentrations of the antibiotics belonging to the classes of β-lactam/cephalosporin/carbapenem (Amoxicillin, Ampicillin, Cefaclor, Cefadroxil, Cefalonium, Cefazolin, Cefoperazone, Cefquinome, Ceftazidime, Ceftiofur, Dicloxacillin, Meropenem, Oxacillin, Penicillin V), macrolides (Azithromycin, Clarithromycin, Erythromycin, Rifaximin, Roxithromycin, Tilmicosin, Tylosin), glycopeptides (Vancomycin), were determined by liquid chromatography-electrospray ionization-quadrupole time of flight-mass spectrometry (LC-ESI-QTOF-MS). All seawater samples were extracted on board using HORIZON SPE-DEX 4790 device (United States). The samples were spiked with internal standards for quality assurance and quality control purposes and were concentrated on Atlantic HLB-M Disk with 47 mm disk holder according to an automated extraction programme (Alygizakis et al., 2019). The extracts were evaporated using nitrogen and reconstituted in methanol:water (50:50 v/v) to the final volume of 500 μL achieving concentration factor of 4,000. The samples were filtered through 0.2 μm RC syringe filter before the LC-ESI-QTOF-MS analysis. The instrumental setup and gradient programme can be found elsewhere (Gago-Ferrero et al., 2020). The method was previously validated for linearity, accuracy using recovery experiments, repeatability and sensitivity (Gago-Ferrero et al., 2020).
Statistical Analysis
The obtained data was analyzed in the RStudio (version 3.6.0, packages: vegan, tidyverse, ade4, geosphere, corrplot, ggplot2). The data on the ARG abundance was tested for the normality by the Shapiro-Wilk normality test, which revealed non-normal distribution (p-value <0.05). Wilcoxon rank sum test was applied (q-value = 0.05), p-value adjustment method: fdr (q-value <0.05), to test for the difference in the genes’ abundance in the water from different Black Sea locations. Spearman correlation was used to test the correlation between ARGs’ abundance and number of the faecal pollution markers. Mantel test was performed with the purpose to infer whether the variance in genes’ number is explained by geographical distance, using the abundance matrix (based on Bray–Curtis measure) and the geographical distance matrix (based on Haversine distance).
Results
Number of Antibiotic Resistance Genes in the Black Sea Water Samples
All target antibiotic resistance genes were found in all Black Sea water samples (Figure 2). Absolute values of the ARGs in each sample are presented in Supplementary Table S2.
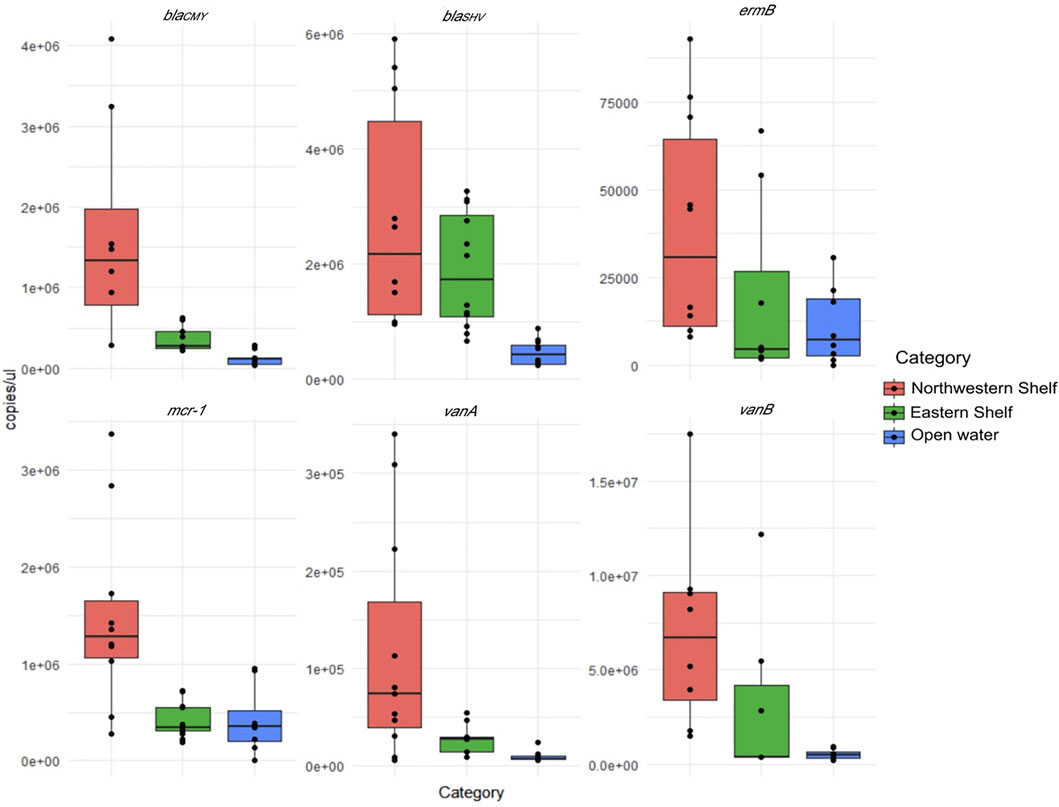
FIGURE 2. Absolute number of ARGs in northwest shelf (stations 1A, 2A, 2, and 3), eastern shelf (stations 9, 10, 11, 12) and open waters (stations 4, 5, 7 and 8).
The average values normalized to the 16S rRNA gene number indicate that the vanB (2 × 10−1 ± 1 × 10−1) was the most numerous gene followed by blaSHV (4 × 10−2 ± 1 × 10−2), mcr-1 (5 × 10−2 ± 3 × 10−2) and blaCMY (1 × 10−2 ± 3 × 10−3). ermB (1 × 10−3 ± 5 × 10−4) and vanA (1 × 10−5 ± 5 × 10−4) were the least numerous genes.
Considering the absolute values, differential abundance of ARGs in the waters of eastern, northwest shelf and open waters was pronounced. The higher number of ARGs in the waters of the northwest shelf was especially notable (Figure 2). Open waters contained the least number of all ARGs. Wilcoxon pairwise comparison proved that blaCMY, vanA and mcr-1 had significantly higher abundance in the northwest shelf coastal area compared to the open waters and the eastern shelf (Wilcoxon, p < 0.05—Table 2). BlaSHV and blaCMY had significantly higher estimates in the eastern shelf compared to the open waters, while blaCMY and mcr-1 significantly varied in their abundance in the eastern and northwestern shelf.
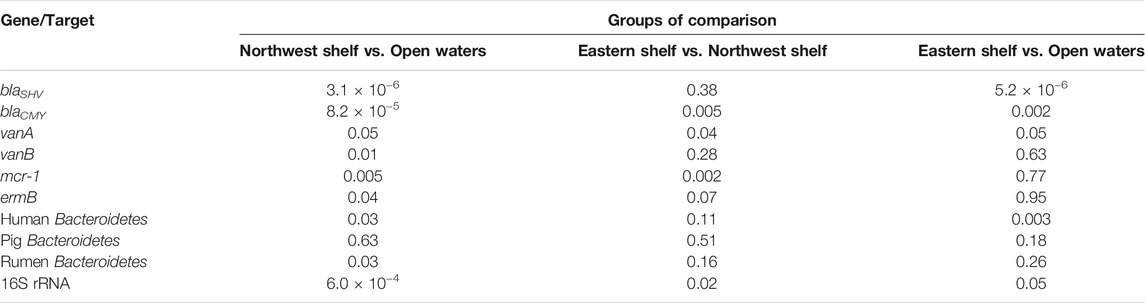
TABLE 2. Wilcoxon rank sum test comparison of the AGS’ absolute number in eastern shelf, open waters, northwest shelf. p-value adjustment method: fdr (q-value<0.05).
The data normalized to the 16S rRNA did not exhibit a pronounced difference in the three Black Sea areas due to the uneven distribution of 16S rRNA gene. It had the highest estimates in the northwest shelf (3 × 108 ± 6 × 107 copies/μL) and the lowest in the open waters (3 × 107 ± 8 × 106 copies/μL). The 16S rRNA copy number was 1×108 ± 5 × 107 copies/μL in the eastern shelf.
Pollution by Faecal Markers
Genetic markers for human-, rumen- and pig-specific faecal members of the Bacteroidetes phylum were quantified to identify the sources of faecal contamination. Anaerobic bacteria belonging to Bacteroidetes phylum inhabit the faeces and exhibit host adaptation on the genetic level (Dick et al., 2005). For this reason Bacteroidetes’ markers (specific regions of the 16S rRNA gene) are used as indicators that can discriminate between human and nonhuman sources of faecal contamination (Reischer et al., 2006; Mieszkin et al., 2009; Epa and of Science, 2019).
Gene sequences of human-associated Bacteroidetes are usually found in the humans’ faeces and environments polluted with human faeces. Human-associated Bacteroidetes can be present in other animals’ sources (Layton et al., 2013). Similarly, pig-associated or cow-associated Bacteroidetes can have cross-reaction in other types of faeces due to the common omnivorous diet and similar digestive tract (Dick et al., 2005; Mieszkin et al., 2009). Nevertheless, presence of human-specific Bacteroidetes in environmental samples is usually associated with human faecal pollution (Layton et al., 2013).
Number of human associated Bacteroidetes (per μL) was 7 × 103 ± 9 × 102 in the northwest shelf, 7 × 103 ± 1 × 102 in the open water and 6 × 103 ± 2 × 103 in the eastern shelf. Number of pig associated Bacteroidetes did not vary in samples taken from different locations, presenting 1 × 102 ± 5 × 101, 2 × 102 ± 1 × 102, 1 × 102 ± 5 × 101 marker copy/μL in the northwest shelf, open waters and eastern shelf respectively. Number of rumen-specific Bacteroidetes was 6 × 102 ± 2 × 102, 4 × 102 ± 4 × 102, 4 × 102 ± 2 × 102 copies/μL in the three respective studied areas.
Human-specific and rumen-specific Bacteroidetes differed in quantity in the northwest coastal waters and open waters according to the Wilcoxon rank sum test (Table 2). Mantel test revealed moderate positive correlation (r = 0.52, p-value = 0.0033) between the faecal markers and physical separation within the northwest shelf—open waters segment (from station 1A to station 8). There was no correlation if the whole distance (from station 1A to station 12) or open water—eastern shelf segment (from station 4 to station 12) was considered (Figure 3). This finding suggests that markers of the faecal pollution mainly enter the Black Sea environment from the northwest coast.
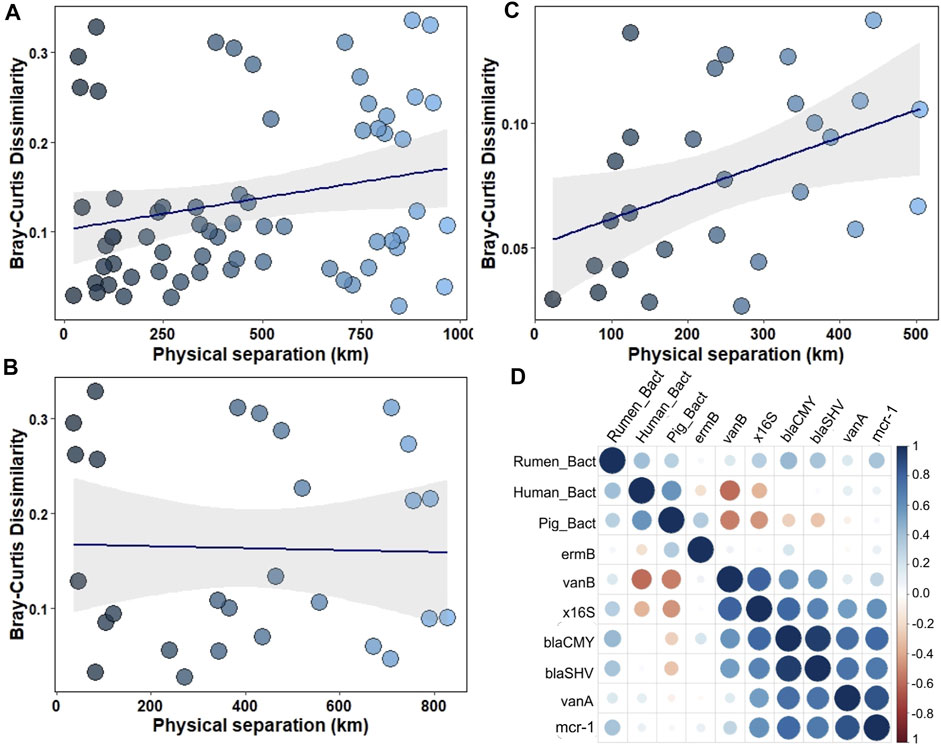
FIGURE 3. (A–C) Variance of faecal pollution dependent on the physical separation of the samples: (A) within the northwest shelf—eastern shelf segment (from station 1A to station 12); (B)—within the open waters—eastern shelf segment (from station 4 to station 12); (C)— within the northwest shelf—open waters segment (from station 1A to station 8); (D)—Spearman correlation between ARGs' number and number of faecal pollution markers.
Rumen-specific Bacteroidetes had moderate positive correlation (0.4, p < 0.05) with blaCMY, blaSHV and mcr-1 genes (Figure 3D). The 16S rRNA gene had strong correlation with blaCMY, blaSHV, vanA and mcr-1 genes (Figure 3D).
Concentrations of Antibiotics
None of the tested antibiotics exceeded the screening detection limit (1.25 ng/L) possibly due to the high dilution of the analytes in seawater.
Discussion
The distribution of ARGs in the Black Sea was evaluated on the sea region-wide scale involving sampling locations in the eastern shelf, open waters and northwest shelf. The list of genes included blaCMY and blaSHV genes that are responsible for the inactivation of beta-lactam antibiotics, ermB targeting macrolides, vanA and vanB genes that inactivate glycopeptides and mcr-1 that compromises effectiveness of colistin.
All studied ARGs were found in the seawater samples. vanB, blaSHV and blaCMY were among the most numerous ARGs. mcr-1 had also high estimates, which is of particular interest since, to our knowledge, this is the first report of the mcr-1 distribution in the Black Sea. Results of the study reveal that vanA and ermB were the least numerous genes among the tested ones, but they were also found in all analysed samples.
The obtained data was compared to ARGs distribution in other aquatic environments summarized in the holistic analysis of the 122 metagenomic sets from lakes and seas (Yang et al., 2019). According to Yang et al. (2019) the relative abundance of beta-lactams’ and aminoglycosides’ resistance genes was in the range from 1 × 10−3 to 2 × 10−2 copies normalized to the 16S rRNA. The copy number of polymyxin and vancomycin resistance genes was on average by one order of magnitude lower (up to 3 × 10−3). Our data on genes coding for aminoglycosides’ resistance is in consistence with that of Yang et al. (2019). The sum of blaSHV and blaCMY varied from 7 × 10−3 to 2 × 10−1 copies that is higher compared to the sum of beta-lactam resistance genes presented by Yang et al. (2019). The copy number of beta-lactam resistance genes (blaOXA) in the Danube River Basin had wider variation compared to our data: 2.4 × 10−8–3.1 × 10−2 (Alygizakis et al., 2019). blaCTXM was detected in the Black Sea water by Sabatino et al. (2020), but its quantity was not estimated.
The present study revealed higher vanB and mcr-1 copy number compared to the estimates of vancomycin and polymixyn resistance genes presented by Yang et al. (2019). According to our data vanB reached 2 × 10−1 ± 1 × 10−1 copies normalized to 16S rRNA in the Black Sea water. This gene was not detected in the effluents of WWTPs in the Danube River Basin, while vanA occurred sporadically (Alygizakis et al., 2019). That indicates that the Danube river is likely not a source of the vanB in the Black Sea, and the other rivers (Dnieper, Dniester or Western Bug) on the northwest site should be considered in this regard. The number of mcr-1 genes varied from 2 × 10−3 to 3 × 10−2 copies normalized to 16S rRNA.
The copy number of ermB (1.8 × 10−5–4.9 × 10−3) in the effluents of WWTPs in the Danube River Basin (Alygizakis et al., 2019) may indicate the origin of this gene distribution. Contrasting with our data, no ermB was detected in the Black Sea water samples according to (Sabatino et al., 2020). This indicates that ermB dissemination in the Black Sea might be possibly on its rise.
High number of ARGs in the Black Sea is likely associated with prescription rates and poorly controlled antibiotic consumption in the riparian countries. Amoxicillin, ceftriaxone and cefuroxime were the most consumed antibiotics during 2013–2018 in Ukraine (Yakovlieva and Bahlai, 2019), which is in parallel with the increase in macrolides consumption in this region (Matyashova and Iakovlieva, 2015). According to (Tarcea Bizo et al., 2015), vancomycin is the most prescribed antibiotic in a university tertiary hospital in Cluj-Napoca, Romania. Meanwhile, European Resistance Surveillance Network (EARS-NET, 2019) reports distribution of hospital isolates resistant to beta-lactams and vancomycin in Black Sea countries. About 63% of E. coli isolates were resistant to amoxicillin/ampicillin in hospitals of Romania and Bulgaria. From 64.1 to 75.7% of K. pneumoniae isolates were resistant to cefotaxime/ceftriaxone/ceftazidime in these two countries respectively. About 35.7% isolates of E. faecium from hospitals of Romania were resistant to vancomycin. Joint Danube Survey 3 results indicate the distribution of the beta-lactam and carbapenem resistant Pseudomonas spp., Klebsiella spp. and E. coli in the Danube water (Kittinger et al., 2016a; Kittinger et al., 2016b). Isolates with resistance patterns normally associated with intensive care units were also found in the Danube river water (Kittinger et al., 2016a). Unfortunately, data reporting the colistin resistance is not presented in the EARS-NET Report. Though, besides the human treatment, colistin is widely applied in veterinary medicine, which fuels the development of the resistance to this antibiotic (Kempf et al., 2016).
The differential abundance of all tested ARGs according to the location of the sampling stations is notable. ARGs were significantly higher (Wilcoxon rank sum test, p < 0.05) in number in the northwest shelf compared to the open waters. blaCMY and blaSHV were considerably more abundant in the eastern shelf than in the open waters. It goes in line with the data on sul1 distribution in the coastal waters of the Black Sea (Sabatino et al., 2020). Similar effect was observed in coastal and open waters of the Bohai Sea and Yellow Sea areas (Lu et al., 2019). The difference in ARG number in the open and coastal waters clearly evidences the anthropogenic impact on the ARGs’ distribution in the Black Sea environment.
The main sources of the coastal and estuarine pollution with ARGs are riverine runoffs, WWTPs, aquaculture and untreated sewage (Zheng et al., 2021). Big cities such as Odessa (Ukraine) and Constanta (Romania) along with a number of smaller towns (total population more than 1.5 million) are located on the northwest bank of the Black Sea. Moreover, Dnieper, Western Bug, Dniester and Danube rivers (total basin area is 1,437,790 km2) fall into the Black Sea on this side providing input of ARGs and other pollutants. Analysis of effluent wastewater from WWTPs in the Danube River Basin has shown the abundance of ARGs (aph(III)a, blaOXA, ermB, tetM etc.,) in the wastewater released into the Danube river and its tributaries (Alygizakis et al., 2019). Similar accumulation of the ARGs is likely to occur in the other rivers. The eastern bank of the Black Sea is less populated with Batumi (Georgia) as the biggest city. Rivers Chorokhi and Rioni fall into the Black Sea on the eastern side and may provide input of the ARGs collected along their basins (total area of basins is 35,500 km2). The higher number of the vanA and mcr-1 in the northwest shelf compared to the eastern shelf is worth noting. Wider area of river basins falling on the north in the Black Sea, extensive agriculture and peculiarities of antibiotic consumption in the region can cause the differences in ARGs’ abundance.
ARGs can persist in the environment due to the presence of antibiotics or disinfectants (Zheng et al., 2021), which provide the selective pressure and facilitate the HGT. On the other hand, faecal pollution is a straightforward source of ARGs dissemination in the environment since gut microbiota is a reservoir of antibiotic-resistant bacteria (Karkman et al., 2018; Khan et al., 2019). The concentrations of 22 targeted antibiotics were below the detection limit (1.25 ng/L) in all tested samples. This excludes the influence of antibiotics on the ARGs proliferation, while the ARGs’ number decrease in the open waters evidences the elimination of the ARGs from the environment.
Previous studies indicated that faecal pollution (from urban areas, animal farms and pasture) was a crucial problem throughout the Danube River Basin (Kirschner et al., 2009). Obviously, the Black Sea is impacted by faecal polluted water from the Danube river and other tributaries, which is reflected in elevated concentration of the markers of faecal pollution in the northwest shelf area. blaCMY, blaSHV and mcr-1 had moderate positive correlation with the Bacteroidetes common to the ruminant microbiome. The positive moderate correlation may evidence the impact of the beta-lactams and colistin use in the veterinary pharmaceuticals on the dissemination of these ARGs in the Black Sea. However, the faecal pollution is likely not the major reason for increased concentration of the ARGs in the coastal waters. Among the other reasons can be pollution with ARBs harbouring the corresponding ARGs since all ARGs had a strong positive correlation with 16S rRNA gene. It can be noted that ARGs could be selected for and transmitted to the environmental bacteria before entering the Black Sea (via WWTPs, sewage, etc.,), while the Black Sea receives the ARBs together with riverine and sewage input.
Conclusion
The distribution of ARGs targeting a wide spectrum of antibiotics (beta-lactams, macrolides, glycopeptides, colistin) including both first-line and last-resort antibiotics was investigated in the Black Sea water samples at a region-wide scale, including shelf and offshore areas. All studied ARGs were detected in all of the studied samples. The number of ARGs inactivating beta-lactams, vancomycin and polymyxin was higher compared to the previous studies performed in the region. The elevated number of colistin resistance mcr-1 gene is of particular interest, as this gene was reported in Black Sea microbial communities for the first time.
ARGs were significantly higher in number in the shelf areas compared to the open waters, which signals involvement of the anthropogenic impact on the natural resistome of the water ecosystem.
The ARGs targeting beta-lactams, vancomycin and last resort antibiotic colistin were of the highest abundance in the northwest shelf, which might be attributed to poorly controlled antibiotic consumption in the riparian countries.
The observed correlation between blaCMY, blaSHV and mcr-1 with the Bacteroidetes common to the ruminant microbiome indicates that the potential sources of ARGs in the Black Sea are the riverine and sewage inflow and, to the lesser extent, application of veterinary pharmaceuticals.
The results are of high environmental concern, highlighting an alarming presence of antibiotic resistance in the Black Sea. An inclusion of ARGs into regular legacy monitoring of the Black Sea and major inflowing rivers is recommended.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
IP: Formal analysis (lead); Investigation (equal); Methodology (equal); Visualization (equal); Writing—original draft (lead). MP: Investigation (equal); Methodology (equal); Validation (equal); Visualization (supporting); Writing—original draft (supporting). AD: Investigation (supporting); Validation (supporting); Visualization (supporting). ED: Investigation (supporting); Project administration (equal); Supervision (equal). NA: Formal analysis (supporting), Methodology (supporting), Writing—original draft (supporting). JS: Formal analysis (supporting); Project administration (equal); Resources (lead); Writing—original draft (supporting).
Funding
This work was supported by the EU/UNDP Project: Improving Environmental Monitoring in the Black Sea—Selected Measures (EMBLAS-Plus), ENPI/2018/389-859.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.823172/full#supplementary-material
References
Alygizakis, N. A., Besselink, H., Paulus, G. K., Oswald, P., Hornstra, L. M., Oswaldova, M., et al. (2019). Characterization of Wastewater Effluents in the Danube River Basin with Chemical Screening, In Vitro Bioassays and Antibiotic Resistant Genes Analysis. Environ. Int. 127, 420–429. doi:10.1016/j.envint.2019.03.060
Amarasiri, M., Sano, D., and Suzuki, S. (2020). Understanding Human Health Risks Caused by Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARG) in Water Environments: Current Knowledge and Questions to Be Answered. Crit. Rev. Environ. Sci. Tech. 50 (19), 2016–2059. doi:10.1080/10643389.2019.1692611
Bakan, G., and Ariman, S. (2004). Persistent Organochlorine Residues in Sediments along the Coast of Mid-black Sea Region of Turkey. Mar. Pollut. Bull. 48 (11), 1031–1039. doi:10.1016/j.marpolbul.2003.12.005
Diamanti, K. S., Alygizakis, N. A., Nika, M.-C., Oswaldova, M., Oswald, P., Thomaidis, N. S., et al. (2020). Assessment of the Chemical Pollution Status of the Dniester River Basin by Wide-Scope Target and Suspect Screening Using Mass Spectrometric Techniques. Anal. Bioanal. Chem. 412 (20), 4893–4907. doi:10.1007/s00216-020-02648-y
Dick, L. K., Bernhard, A. E., Brodeur, T. J., Santo Domingo, J. W., Simpson, J. M., Walters, S. P., et al. (2005). Host Distributions of Uncultivated Fecal Bacteroidales Bacteria Reveal Genetic Markers for Fecal Source Identification. Appl. Environ. Microbiol. 71 (6), 3184–3191. doi:10.1128/AEM.71.6.3184-3191.2005
Epa and of Science (2019). Office of Water Method 1697: Characterization of Human Fecal Pollution in Water by HF183/BacR287 TaqMan ® Quantitative Polymerase Chain Reaction (qPCR) Assay I. EPA Off. Water 821, 2–19. Marchwww.epa.gov.
Gago-Ferrero, P., Bletsou, A. A., Damalas, D. E., Aalizadeh, R., Alygizakis, N. A., Singer, H. P., et al. (2020). Wide-scope Target Screening of >2000 Emerging Contaminants in Wastewater Samples with UPLC-Q-ToF-HRMS/MS and Smart Evaluation of its Performance through the Validation of 195 Selected Representative Analytes. J. Hazard. Mater. 387, 121712. doi:10.1016/j.jhazmat.2019.121712
Griffin, D. W., Benzel, W. M., Fisher, S. C., Focazio, M. J., Iwanowicz, L. R., Loftin, K. A., et al. (2019). The Presence of Antibiotic Resistance Genes in Coastal Soil and Sediment Samples from the Eastern Seaboard of the USA. Environ. Monit. Assess. 191 (Suppl. 2), 300. doi:10.1007/s10661-019-7426-z
Hembach, N., Schmid, F., Alexander, J., Hiller, C., Rogall, E. T., and Schwartz, T. (2017). Occurrence of the Mcr-1 Colistin Resistance Gene and Other Clinically Relevant Antibiotic Resistance Genes in Microbial Populations at Different Municipal Wastewater Treatment Plants in Germany. Front. Microbiol. 8, 1282. doi:10.3389/fmicb.2017.01282
Karkman, A., Do, T. T., Walsh, F., and Virta, M. P. J. (2018). Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 26 (3), 220–228. doi:10.1016/j.tim.2017.09.005
Karkman, A., Pärnänen, K., and Larsson, D. G. J. (2019). Fecal Pollution Can Explain Antibiotic Resistance Gene Abundances in Anthropogenically Impacted Environments. Nat. Commun. 10 (1), 80. doi:10.1038/s41467-018-07992-3
Kayis, S., Capkin, E., and Altinok, Ii. (2009). Bacteria in Rainbow Trout (Oncorhynchus mykiss) in the Southern Black Sea Region of Turkey - A Survey. Israeli J. Aquacult. - Bamidgeh 61 (4), 339–344. http://hdl.handle.net/10524/19300.
Kayiş, Ş., Soyköse, G., İpek, Z. Z., and Er, A. (2021). Determination of Bacterial Contamination and Antibiotic Resistance of the Bacteria in the Some Trout Farm Hatcheries in the Eastern Black Sea Region of Turkey. J. Limnology Freshw. Fish. Res. 7 (2), 101–107. doi:10.17216/limnofish.827718
Kempf, I., Jouy, E., and Chauvin, C. (2016). Colistin Use and Colistin Resistance in Bacteria from Animals. Int. J. Antimicrob. Agents 48 (6), 598–606. doi:10.1016/j.ijantimicag.2016.09.016
Khan, F. A., Söderquist, B., and Jass, J. (2019). Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front. Microbiol. 10, 688. doi:10.3389/fmicb.2019.00688
Kirschner, A. K. T., Kavka, G. G., Velimirov, B., Mach, R. L., Sommer, R., and Farnleitner, A. H. (2009). Microbiological Water Quality along the Danube River: Integrating Data from Two Whole-River Surveys and a Transnational Monitoring Network. Water Res. 43 (15), 3673–3684. doi:10.1016/j.watres.2009.05.034
Kittinger, C., Lipp, M., Baumert, R., Folli, B., Koraimann, G., Toplitsch, D., et al. (2016a). Antibiotic Resistance Patterns of Pseudomonas Spp. Isolated from the River Danube. Front. Microbiol. 7, 586. doi:10.3389/fmicb.2016.00586
Kittinger, C., Lipp, M., Folli, B., Kirschner, A., Baumert, R., Galler, H., et al. (2016b). Enterobacteriaceae Isolated from the River Danube: Antibiotic Resistances, with a Focus on the Presence of ESBL and Carbapenemases. PLOS ONE 11 (11), e0165820. doi:10.1371/journal.pone.0165820
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 41, e1. doi:10.1093/nar/gks808
Kraemer, S. A., Ramachandran, A., and Perron, G. G. (2019). Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 7 (6), 180. doi:10.3390/microorganisms7060180
Kümmerer, K. (2009). Antibiotics in the Aquatic Environment - A Review - Part I. Chemosphere 75 (4), 417–434. doi:10.1016/j.chemosphere.2008.11.086
Layton, B. A., Cao, Y., Ebentier, D. L., Hanley, K., Ballesté, E., Brandão, J., et al. (2013). Performance of Human Fecal Anaerobe-Associated PCR-Based Assays in a Multi-Laboratory Method Evaluation Study. Water Res. 47 (18), 6897–6908. doi:10.1016/j.watres.2013.05.060
Lu, J., Zhang, Y., Wu, J., Wang, J., Zhang, C., and Lin, Y. (2019). Occurrence and Spatial Distribution of Antibiotic Resistance Genes in the Bohai Sea and Yellow Sea Areas, China. Environ. Pollut. 252 (Pt A), 450–460. doi:10.1016/j.envpol.2019.05.143
Matyashova, N. O., and Iakovlieva, L. V. (2015). Analysis of Consumption of Macrolides in Ukraine. Klìn. Farm. 19 (2), 19–22. doi:10.24959/cphj.15.1338
Mieszkin, S., Furet, J.-P., Corthier, G., and Gourmelon, M. (2009). Estimation of Pig Fecal Contamination in a River Catchment by Real-Time PCR Using Two Pig-specific Bacteroidales 16S rRNA Genetic Markers. Appl. Environ. Microbiol. 75 (10), 3045–3054. doi:10.1128/AEM.02343-08
Mirzaei, B., Babaei, R., Asiabar, A. P. D., and Bameri, Z. (2015). Detection of Both vanA & vanB Genes in vanA Phenotypes of Enterococci by Taq Man RT-PCR. Braz. J. Microbiol.[Publication Braz. Soc. Microbiol. 46 (1), 161–165. doi:10.1590/S1517-838246120131234
Ozkoc, H. B., Bakan, G., and Ariman, S. (2007). Distribution and Bioaccumulation of Organochlorine Pesticides along the Black Sea Coast. Environ. Geochem. Health 29 (1), 59–68. doi:10.1007/s10653-006-9064-y
Reischer, G. H., Kasper, D. C., Steinborn, R., Mach, R. L., and Farnleitner, A. H. (2006). Quantitative PCR Method for Sensitive Detection of Ruminant Fecal Pollution in Freshwater and Evaluation of This Method in alpine Karstic Regions. Appl. Environ. Microbiol. 72 (8), 5610–5614. doi:10.1128/AEM.00364-06
Roschanski, N., Fischer, J., Guerra, B., and Roesler, U. (2014). Development of a Multiplex Real-Time PCR for the Rapid Detection of the Predominant Beta-Lactamase Genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLOS ONE 9 (7), e100956. doi:10.1371/journal.pone.0100956
Sabatino, R., Di Cesare, A., Dzhembekova, N., Fontaneto, D., Eckert, E. M., Corno, G., et al. (2020). Spatial Distribution of Antibiotic and Heavy Metal Resistance Genes in the Black Sea. Mar. Pollut. Bull. 160, 111635. doi:10.1016/j.marpolbul.2020.111635
Shimkus, K. M., and Trimonis, E. S. (1974). “Modern Sedimentation in Black Sea1,” in The Black Sea—Geology, Chemistry, and Biology. Editors E. T. Degens, and D. A. Ross (American Association of Petroleum Geologists), 20, 0. doi:10.1306/M20377C23
Stoichev, T., Makedonski, L., Trifonova, T., Stancheva, M., and Ribarova, F. (2007). DDT in Fish from the Bulgarian Region of the Black Sea. Chem. Ecol. 23 (3), 191–200. doi:10.1080/02757540701339851
Szczepanowski, R., Linke, B., Krahn, I., Gartemann, K.-H., Gützkow, T., Eichler, W., et al. (2009). Detection of 140 Clinically Relevant Antibiotic-Resistance Genes in the Plasmid Metagenome of Wastewater Treatment Plant Bacteria Showing Reduced Susceptibility to Selected Antibiotics. Microbiology (Reading, England) 155 (Pt 7), 2306–2319. doi:10.1099/mic.0.028233-0
Szekeres, E., Chiriac, C. M., Baricz, A., Szőke-Nagy, T., Lung, I., Soran, M.-L., et al. (2018). Investigating Antibiotics, Antibiotic Resistance Genes, and Microbial Contaminants in Groundwater in Relation to the Proximity of Urban Areas. Environ. Pollut. 236, 734–744. doi:10.1016/j.envpol.2018.01.107
Tang, J., Shi, T., Wu, X., Cao, H., Li, X., Hua, R., et al. (2015). The Occurrence and Distribution of Antibiotics in Lake Chaohu, China: Seasonal Variation, Potential Source and Risk Assessment. Chemosphere 122, 154–161. doi:10.1016/j.chemosphere.2014.11.032
Tarcea Bizo, P., Dumitras, D., and Popa, A. (2015). Evaluation of Restricted Antibiotic Use in a Hospital in Romania. Int. J. Clin. Pharm. 37 (3), 452–456. doi:10.1007/s11096-015-0096-1
Terzi, E., and Isler, H. (2019). Antibiotic Resistance Genes of Escherichia coli in Coastal Marine Environment of Eastern Black Sea, Turkey. Fresenius Environ. Bull. 28 (2A). https://hdl.handle.net/11436/1671.
Türe, M., and Alp, H. (2016). Identification of Bacterial Pathogens and Determination of Their Antibacterial Resistance Profiles in Some Cultured Fish in Turkey. J. Vet. Res. 60 (2), 141–146. doi:10.1515/jvetres-2016-0020
Yakovlieva, L., and Bahlai, T. (2019). Β-Lactam Antibiotics in Ukraine: Market and Consumption Analysis in 2013-2018. ScienceRise: Pharm. Sci. 2 (18), 16–21. doi:10.15587/2519-4852.2019.165682
Yang, Y., Li, Z., Song, W., Du, L., Ye, C., Zhao, B., et al. (2019). Metagenomic Insights into the Abundance and Composition of Resistance Genes in Aquatic Environments: Influence of Stratification and Geography. Environ. Int. 127, 371–380. doi:10.1016/j.envint.2019.03.062
Zarfel, G., Folli, B., Lipp, M., Pfeifer, B., Baumert, R., Farnleitner, A., et al. (2015). “Spread of Non-wild Type Antibiotic Resistant Phenotypes in the River Danube,” in Joint Danube Survey 3: A Comprehensive Analysis of Danube Water QualityICPDR – International Commission for the Protection of the Danube River. Editors I. Liška, F. Wagner, M. Sengl, K. Deutsch, and J. Slobodník, 169–172. http://www.danubesurvey.org/jds3/jds3-files/nodes/documents/jds3_final_scientific_report_1.pdf.
Keywords: ARGs, beta-lactam, macrolide, colistin, vancomycin, qPCR, Black Sea
Citation: Prekrasna I, Pavlovska M, Dzhulai A, Dykyi E, Alygizakis N and Slobodnik J (2022) Antibiotic Resistance in Black Sea Microbial Communities. Front. Environ. Sci. 10:823172. doi: 10.3389/fenvs.2022.823172
Received: 26 November 2021; Accepted: 24 January 2022;
Published: 21 February 2022.
Edited by:
Marcelo Pedrosa Gomes, Federal University of Paraná, BrazilReviewed by:
Yi-Yun Liu, South China Agricultural University, ChinaYuyi Yang, Wuhan Botanical Garden (CAS), China
Copyright © 2022 Prekrasna, Pavlovska, Dzhulai, Dykyi, Alygizakis and Slobodnik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaroslav Slobodnik, slobodnik@ei.sk
 Ievgeniia Prekrasna
Ievgeniia Prekrasna Mariia Pavlovska
Mariia Pavlovska Artem Dzhulai
Artem Dzhulai Evgen Dykyi
Evgen Dykyi Nikiforos Alygizakis
Nikiforos Alygizakis Jaroslav Slobodnik3*
Jaroslav Slobodnik3*