Pangeo Forge: Crowdsourcing Analysis-Ready, Cloud Optimized Data Production
- 1Lamont-Doherty Earth Observatory, Columbia University, Palisades, NY, United States
- 2CarbonPlan, San Francisco, CA, United States
- 3National Center for Atmospheric Research, Boulder, CO, United States
- 4Department of Atmospheric and Ocean Science, University of Maryland, College Park, MD, United States
- 5Development Seed, Washington, DC, United States
- 6Google Research, Mountain View, CA, United States
Pangeo Forge is a new community-driven platform that accelerates science by providing high-level recipe frameworks alongside cloud compute infrastructure for extracting data from provider archives, transforming it into analysis-ready, cloud-optimized (ARCO) data stores, and providing a human- and machine-readable catalog for browsing and loading. In abstracting the scientific domain logic of data recipes from cloud infrastructure concerns, Pangeo Forge aims to open a door for a broader community of scientists to participate in ARCO data production. A wholly open-source platform composed of multiple modular components, Pangeo Forge presents a foundation for the practice of reproducible, cloud-native, big-data ocean, weather, and climate science without relying on proprietary or cloud-vendor-specific tooling.
1. Introduction
In the past 10 years, we have witnessed a rapid transformation in environmental data access and analysis. The old paradigm, which we refer to as the download model, was to search for files from a range of different data providers, download them to a local laptop or workstation, and analyze the data in a traditional desktop-based analysis environment (e.g., IDL, MATLAB, and ArcGIS). The new paradigm, which we call data-proximate computing, instead brings compute resources adjacent to the data, with users performing their data analysis in a web browser and retrieving data on demand via APIs or HTTP calls (Ramamurthy, 2017). Data-proximate environmental data analysis tools and platforms are often deployed in the commercial cloud, which provides scalable, on-demand computing and high-throughput data access, but are not necessarily limited to cloud environments. Data-proximate computing removes the burden on the data user to provide local computing; this has the potential to massively expand access to environmental data, empowering communities that have been historically marginalized and lack such local computing resources (Gentemann et al., 2021). However, this democratization is not guaranteed. FAIR data, open standards, and equitable access to resources must be actively pursued by the community (Wilkinson et al., 2016; Stall et al., 2019).
Many different platforms exist to analyze environmental data in the cloud; e.g., Google Earth Engine (GEE) and Microsoft's Planetary Computer (Gorelick et al., 2017; Microsoft, 2021). A common need for all such platforms is access to analysis-ready, cloud optimized (ARCO) data. While a range of powerful ARCO data formats exist (e.g., Cloud Optimized GeoTIFF, Zarr, TileDB Embedded, and Parquet), ARCO data production has remained a bespoke, labor-intensive process. Recent sessions devoted to cloud computing at meetings of the American Geophysical Union (AGU) and Earth System Information Partners (ESIP) enumerated the considerable toil involved in creating ARCO data in the cloud (Hua et al., 2020; Quinn et al., 2020). For example, when GEE partnered with the European Center for Medium-Range Weather Forecasting (ECMWF) to bring a portion of the ERA5 reanalysis data to GEE, the data ingestion process was incredibly time and resource intensive, spanning 9 months and involving a suite of specialized tools (Wagemann, 2020).
In addition to demanding computing resources and specialized software, ARCO data production also requires knowledge in a range of areas, including: legacy and ARCO data formats, metadata standards, cloud computing APIs, distributed computing frameworks, and domain-specific knowledge sufficient to perform quality control on a particular dataset. In our experience, the number of individuals with this combination of experience is very small, limiting the rate of ARCO data production overall.
This paper describes Pangeo Forge, a new platform for the production of ARCO data (Pangeo Forge Community, 2021). A central goal of Pangeo Forge is to reduce the toil associated with downloading, cleaning, and preparing data for analysis, particularly for the large, complex datasets associated with high-bandwidth observing systems, Earth-system simulations, and weather reanalyses. Recognizing that individuals with domain-specific data knowledge are not necessarily experts in cloud computing or distributed data processing, Pangeo Forge aims to lower the barrier for these scientists to contribute to ARCO data curation. Finally, we hope to build a platform that encourages open and inclusive participation, crowdsourcing ARCO data production from the diverse community of environmental data specialists across the world, for the mutual benefit of all.
At the time of writing, Pangeo Forge is still a work on progress. This paper describes the motivation and inspiration for building the platform (section 2) and reviews its technical design and implementation (section 3). We then describe some example datasets that have been produced with Pangeo Forge (section 4) and conclude with the future outlook for the platform (section 5).
2. Motivation and Inspiration
2.1. Analysis-Ready, Cloud-Optimized Data
In the context of geospatial imagery, remote sensing instruments collect raw data which typically requires preprocessing, including color correction and orthorectification, before being used for analysis. The term analysis-ready data (ARD) emerged originally in this domain, to refer to a temporal stack of satellite images depicting a specific spatial extent and delivered to the end-user or customer with these preprocessing steps applied (Dwyer et al., 2018; Holmes, 2018). In the context of this paper, however, we use the term “analysis-ready” more generally to refer to any dataset that has been preprocessed such that it fulfills the quality standards required by the analysis which will be performed on it. This may include merging and alignment of many individual source files or file-like objects into a single cohesive entity. For remotely sensed measurements, it may involve signal processing to correct for known atmospheric or other distortions. For synthetic (i.e., simulation) data, quality control may include ensuring that output values fall within test parameters defined by the model developers, as well as homogenization of metadata across simulation ensembles.
Analysis-ready data is not necessarily or always cloud-optimized. One way of understanding this is to observe that just because an algorithm can be applied to a given dataset, that fact alone does not guarantee the algorithm will execute expediently or efficiently. In a context where even efficient algorithms can take hours or days to run, optimization matters. Computational performance is affected by many factors including algorithm design and hardware specifications, but in the case of big data analytics, the rate-limiting aspect of the system is often I/O throughput, i.e., the rate at which bytes can be read into the algorithm from the data storage location (Abernathey et al., 2021). This rate is itself influenced by variables such as network bandwidth, hardware characteristics, and data format. When we refer to “cloud-optimized” data it is this third variable, format, which we are most concerned with. Cloud-optimized data formats are unique insofar as they support direct access to data subsets without the computational overhead of opening and navigating through a massive data object simply to retrieve a small subset of bytes within it. Implementations of this functionality vary according to the specific cloud-optimized format: some formats include a metadata header which maps byte-ranges within a single large data object, while others opt to split a large object up into many small blocks stored in an organized hierarchical structure. Regardless of the specific implementation, the end result is an interface whereby algorithms can efficiently access data subsets. Efficient access to data subsets is especially impactful in the context of cloud object storage, where simultaneous read/write of arbitrary numbers of data subsets does not decrease the throughput to any individual subset. As such, parallel I/O dramatically increases cumulative throughput.
Analysis-ready, cloud-optimized datasets are, therefore, datasets which have undergone the preprocessing required to fulfill the quality standards of a particular analytic task and which are also stored in formats that allow efficient, direct access to data subsets.
2.2. Open Science, Open Source
The Pangeo Forge codebase, which is written in Python, is entirely open source, as are its Python dependencies including packages such as NumPy, Xarray, Dask, Filesystem Spec, and Zarr (Dask Development Team, 2016; Hoyer and Hamman, 2017; Harris et al., 2020; Durant, 2021; Miles et al., 2021). We see open source software as a scientific imperative. Production of ARCO datasets involves considerable preprocessing and reformatting. Data corruptions can easily be introduced at any step of these multi-stage transformations, either due to user error or, less commonly but more consequentially, due to bugs in the software packages used to perform the ARCO transformation. In an open source context, the scientific user community can readily introspect every step of the process, building trust in its effectiveness as well as contributing to its robustness by identifying bugs when they arise. The core scientific tenet of reproducibility is also served by open source: the exact provenance of each byte of data that passes through Pangeo Forge is entirely transparent, traceable, and recreatable.
Where Pangeo Forge must unavoidably rely on commercial technology providers, we strive always to uphold the user's Right to Replicate (2i2c.org, 2021). In practice, this means that even if an underlying cloud provider technology is closed source, the application code defining our particular implementation of that technology is always open source, allowing anyone the option to replicate our system exactly as we've deployed it. Version control hosting, continuous integration, compute infrastructure, storage resources, and workflow automation are arenas in which commercial solutions are implemented. The former two services are provided through GitHub repositories and GitHub Actions, respectively, and the latter three through the “big three” cloud service providers (Google Cloud, Amazon Web Services, Microsoft Azure) and Prefect, a dataflow automation provider.
2.3. Crowdsourcing Complexity: the Conda Forge Model
The incredible diversity of environmental science datasets and use cases means that a fully generalized and automatic approach for transforming archival data into ARCO stores is likely neither achievable nor desirable. Depending on the analysis being performed, for example, two users may want the same archival source data in ARCO form, but with different chunking strategies (Chunking, i.e., the internal arrangement of a dataset's bytes, is often adjusted to optimize for different analytical tasks). Transforming just a single dataset from its archival source into an ARCO data store is an incredibly complex task which unavoidably requires human expertise to ensure the result is fit for the intended scientific purpose. Fantasies of cookie-cutter algorithms automatically performing these transformations without human calibration are quickly dispelled by the realities of just how unruly archival data often are, and how purpose-built the ARCO data stores created from them must be. As with all of science, ARCO transformations require human interpretation and judgement.
The necessity of human participation, combined with the exponentially increasing volumes of data being archived, means that ARCO data production is more work than any individual lab, institution, or even federation of institutions could ever aspire to manage in a top-down manner. Any effort to truly address the present scarcity of high-quality ARCO data must by necessity be a grassroots undertaking by the international community of scientists, analysts, and engineers who struggle with these problems on a daily basis.
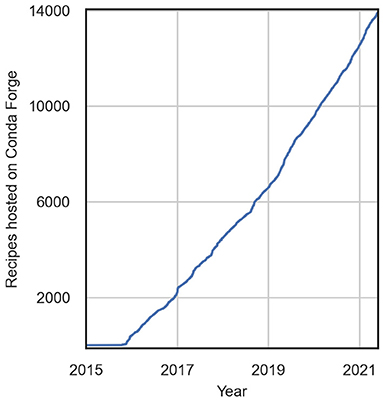
The software packaging utility Conda Forge, from which Pangeo Forge draws both inspiration and its name, provides a successful example of solving a similar problem via crowdsourcing (Conda-Forge Community, 2015). Conda Forge emerged in 2015 in response to frustrations scientific software users consistently faced when attempting to install system package dependencies in the course of their research. Just like ARCO data production, installing open source software packages with binary dependencies is frequently a multi-step process involving an intricate sequence of software compilation. If any one step is completed out of order, or perhaps if one of the sub-packages installed is of the wrong version, the end result will be non-functional. This struggle devoured countless years worth of human effort on the part of researchers who required a specific software configuration to pursue their investigations.
Conda Forge introduced the simple yet revolutionary notion that two people, let alone hundreds or thousands, should not be duplicating effort to accomplish the same tedious tasks. As an alternative to that toil, Conda Forge established a publicly-licensed and freely-accessible storehouse, hosted on the open internet, to hold blueprints for performing these arcane yet essential engineering feats. It also defined a process for contributing blueprints to that storehouse and established a build system compatible with the Conda package manager, a component of the open-source Anaconda Software Distribution, itself a popular collection of data science tooling (Anaconda Inc., 2021). This interconnection with the Conda package manager, in addition to serving as the inspiration for Conda Forge's name, means that a given Conda Forge package can be built from the public storehouse onto a community member's system with just a one-line command: conda install.
It is not an understatement to say that this simple invocation, conda install, and the system built by Anaconda undergirding it, fundamentally transformed for the better the practice of computational science with open source software. The crowdsourcing model defined by Conda Forge then leveraged this technology to maximal advantage for the open source scientific community. For evidence of this fact, we need look no further than the incredible growth rate of community contributed “recipes” (as these installation blueprints are known) in the Conda Forge storehouse (Figure 1). The summed impact of this solution totals untold numbers of reclaimed hours which are now dedicated to scientific research itself, rather than tinkering with finicky engineering issues.
In the case of Conda Forge, community members contribute recipes to a public storehouse which define steps for building software dependencies. Then they, along with anyone else, can avoid ever needing to revisit the toil and time of manually building that specific piece of software again. Contributions to Conda Forge, while they often include executable software components, consist minimally of a single metadata file, named meta.yaml, which conforms to a specification established in accordance with the build system. This design is explicitly copied in Pangeo Forge.
3. Technical Design and Implementation of Pangeo Forge
Pangeo Forge follows an agile development model, characterized by rapid iteration, frequent releases, and continuous feedback from users. As such, implementation details will likely change over time. The following describes the system at the time of publication.
At the highest level, Pangeo Forge consists of three primary components:
• pangeo-forge-recipes: A standalone Python package which provides a data model (“recipes”) and scalable algorithms for ARCO data production. This package can be used by itself, without the platform's cloud automation tools.
• An automation system which executes recipes using distributed processing in the cloud.
• A catalog which exposes the ARCO data to end users.
3.1. Recipes: Object-Oriented Extraction, Optimization, and Storage (EOS) Algorithms
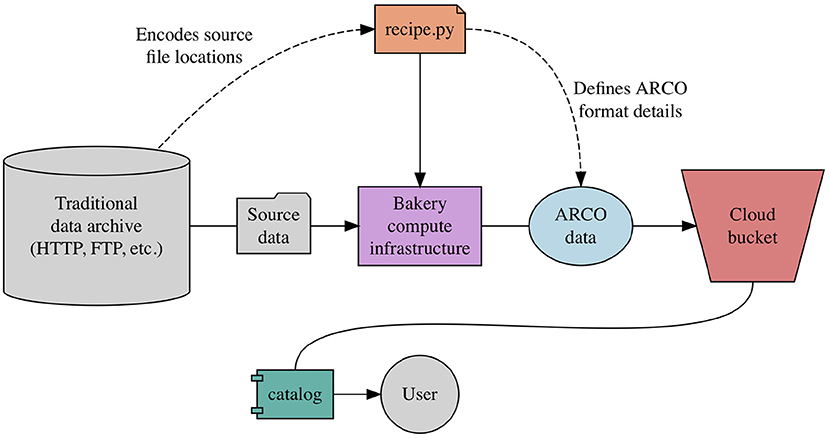
Inspired directly by Conda Forge, Pangeo Forge defines the concept of a recipe, which specifies the logic for transforming a specific data archive into an ARCO data store. All contributions to Pangeo Forge must include an executable Python module, named recipe.py or similar, in which the data transformation logic is embedded (Figure 2). The recipe contributor is expected to use one of a predefined set of template algorithms defined by Pangeo Forge. Each of these templated algorithms is designed to transform data of a particular source type into a corresponding ARCO format, and requires only that the contributor populate the template with information unique to their specific data transformation, including the location of the source files and the way in which they should be aligned in the resulting ARCO data store.
Pangeo Forge implements template algorithms with object-oriented programming (OOP), the predominant style of software design employed in Python software packages. In this style, generic concepts are represented as abstract classes which gain meaning once instantiated with details relevant to a particular use case. Once instantiated, class instances (as they are known) can perform operations on or with the attributes (i.e., details) they ve been given. In Pangeo Forge, the operations embedded in the template algorithms are, broadly speaking, those of data extraction, optimization, storage (EOS). First, data is extracted from a traditional source file server, most commonly via HTTP or FTP request; next, the source data is transformed into an ARCO format; and finally, the data is deposited into cloud object storage.
Within a given class of these EOS algorithms, it's possible to largely generalize the esoteric transformation logic itself, while leaving the specific attributes, such as source file location and alignment criteria, up to the recipe contributor to fill in. The completed recipe.py module containing a specific instance of the generic EOS algorithm can then be executed in one of a number of ways. While recipe developers are certainly free to run these open source algorithms on private compute clusters, they are strongly encouraged to submit their recipes to be run on Pangeo Forge's shared infrastructure, which has the dual benefit of being a freely accessible resource and, perhaps even more importantly, results in the ARCO data being written to a publicly-accessible cloud storage bucket and added to the Pangeo Forge catalog for discovery and shared use by the global community. It is through scaling contributions to our public ARCO data catalog that Pangeo Forge aspires to do for ARCO data production what Conda Forge has already accomplished for software dependency management.
3.2. Base Abstractions: Insulating Scientific Domain Expertise From Cloud Automation Concerns
Pangeo Forge consists of multiple interrelated, modular components. Each of these components, such as the recipes described above, consists of some abstracted notions about how a given aspect of the system typically functions. These abstractions are for the most part implemented as Python classes. They include classes related to source file location, organization, and access requirements; the recipe classes themselves; classes which define storage targets (both for depositing the eventual ARCO data store, as well as for intermediate caching); and multiple different models according to which the algorithms themselves can be executed.
The boundaries between these abstraction categories have been carefully considered with the aim of insulating scientific domain expertise (i.e., of the recipe contributor) from the equally rigorous yet wholly distinct arena of distributed computing and cloud automation. Among ocean, weather, and climate scientists today, Python is a common skill, but the ability to script advanced data analyses by no means guarantees an equivalent fluency in cloud infrastructure deployments, storage interfaces, and workflow engines. Moreover, Pangeo Forge aims to transform entire global datasets, the size of which is often measured in terabytes or petabytes. This scale introduces additional technical challenges and tools which are more specialized than the skills required to convert a small subset of data.
By abstracting data sourcing and quality control (i.e., the recipe domain) from cloud deployment and workflow concerns, Pangeo Forge allows recipe contributors to focus exclusively on defining source file information along with setting parameters for one of the predefined recipe classes. Recipe contributors are, importantly, not expected to understand or manipulate the storage and execution aspects of the system, which are maintained by community members with expertise in those areas. In what follows, well examine four aspects of the system in closer detail.
3.2.1. Source File Patterns
In Pangeo Forge, all data transformations begin with a FilePattern. This Python class encodes information about archival source files including their location, access requirements, and alignment criteria. Data providers such as NASA and NOAA commonly distribute source files over HTTP. File Transfer Protocol (FTP) is also a common means for distribution of source data in the earth and atmospheric sciences. In either case, contributors specify the access URLs for their source files as part of a FilePattern. If the archival data URLs correspond to a dynamic API such as OPeNDAP (Cornillon et al., 2009; Hankin et al., 2010), rather than a static file server, that information is specified at this stage. In cases where authorization credentials such as a password or API token are required to access the source data, the names of environment variables which will point to these values at runtime are included here as well.
Almost all ARCO datasets are assembled from many source files which are typically divided by data providers according to temporal, spatial, and/or variable extents. In addition to defining the location(s) of the source files, the FilePattern is where contributors define how the specified set of source files should be aligned to create a single cohesive ARCO dataset. Alignment operations include concatenation, for arranging files end-to-end; and merging, for layering files which cover the same spatial or temporal extent, but for different variables. Listing 1 demonstrates how a recipe contributor would define a FilePattern for archival data accessed via the imaginary file server http://data-provider.org/. The pattern defined in the final line of this snippet encodes not just the location of the source files, but also the fact that any resulting ARCO data store should concatenate these files in the time dimension, and merge them in the variable dimension. This encoding relies on the near-universal practice among data providers of defining URL naming schemes which are descriptive of a given file server's contents; i.e., the access endpoint for a file covering specific extents will name those extents as part of its URL. The objects merge_dim and concat_dim, in the example provided in Listing 2, map our imaginary file server's URL character string representation of dataset dimensions onto Pangeo Forge internal datatypes for consumption by downstream recipe classes.
3.2.2. Recipe Classes
Ocean, climate, and weather data is archived in a wide range of formats. The core abstractions of Pangeo Forge, including FilePattern, are designed to be agnostic to data formats, and can be leveraged to transform any archival source file format into any corresponding ARCO format. The transformation from a specific archival format (or category of formats) into a corresponding ARCO format does require a dedicated algorithm, however. In Pangeo Forge, recipe classes are the modular template algorithms which perform a specific category of ARCO transformation. As modular components, an arbitrary number of these classes can be added to the platform over time, with each new class adding support for a new type of ARCO data production.
As of the writing of this paper, Pangeo Forge defines two such recipe classes, XarrayZarrRecipe and HDFReferenceRecipe, each of which is most commonly used to transform one or many NetCDF files into a single consolidated Zarr dataset. The difference between these algorithms lies in the nature of their outputs. Whereas, XarrayZarrRecipe creates an actual Zarr store by mirroring the source file bytes into a new format, HDFReferenceRecipe leverages the Python library kerchunk to write lightweight metadata files which map the location of bytes within the archival source files, allowing users to read the original data in a cloud-optimized manner with the Zarr library, but without duplicating bytes (Durant et al., 2021).
As an algorithm case study, we ll take a closer look at the internals of the XarrayZarrRecipe. To begin, let's consider how we would create an instance of this algorithm. In the simplest case each algorithm instance requires only a FilePattern instance as input. Using the instance we defined in Listing 1, we define a recipe as shown in Listing 2. In just these few simple lines, we have created an algorithm containing all of the information needed to extract data from our specified provider archive and transform it into the cloud-optimized Zarr format.
Real-world use cases will likely necessitate additional options be specified for the XarrayZarrRecipe instance. Pangeo Forge supports many such options. One worth highlighting is the target_chunks option, which is used to indicate the desired chunking scheme of the resulting ARCO data store. As mentioned in section 2.3, chunking, the internal subsetting of a large dataset, is often optimized for a particular analytical aim, with a classic example being the divergent chunking required for optimizing timeseries vs. spatial analyses. Contributors pass a mapping of a dimension name to an integer value to specify their desired chunking; e.g., target_chunks={~time~: 10} tells the algorithm to divide the ARCO dataset into chunks of length 10 in the time dimension. Should downstream data users require a variation on this or another contributor-defined option, they can make or request changes to the recipe and release those changes as a new dataset version (see section 3.3.2 for further discussion of dataset versioning).
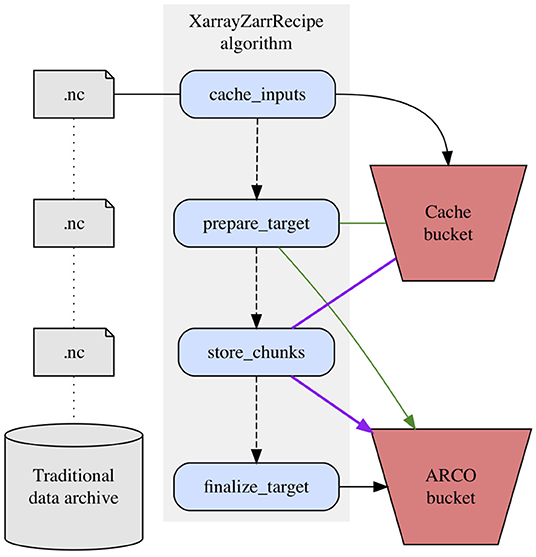
A full treatment of the Zarr specification is beyond the scope of this paper, but a brief overview will provide a better context for understanding. In a Zarr store, compressed chunks of data are stored as individual objects within a hierarchy that includes a single, consolidated JSON metadata file. In actuality, cloud object stores do not implement files and folders, but in a colloquial sense we can imagine a Zarr store as a directory containing a single metadata file alongside arbitrary numbers of data files, each of which contains a chunk of the overall dataset (Miles et al., 2021). The XarrayZarrRecipe algorithm which transforms archival data into this format consists of four sequential steps, each of which performs a series of sub-operations. Depending on the specific use case, one or more of these steps may be omitted, but we will consider them here for the scenario in which they are all performed (Figure 3).
Caching input files is the first step of the XarrayZarrRecipe algorithm. This step copies all archival files required for the dataset into temporary storage in a cloud storage bucket. This affords downstream steps of the algorithm fast, parallelizable access to the source data. Typically, the cached source files will be in NetCDF format (Rew et al., 2006). As the name of the algorithm suggests, however, the actual requirement is not for NetCDF inputs specifically, but rather for input files compatible with Xarray, a widely-used Python interface for labeled multidimensional arrays that supports multiple backend file formats, including GRIB, COG, and some flavors of HDF5 (Hoyer and Hamman, 2017).
Before any actual bytes are written to the Zarr store, the target storage location must first be initialized with the skeletal structure of the ARCO dataset. We refer to this step, which immediately follows caching, as prepare_target. Preparing the target entails reading metadata from a representative subset of the source files to establish an empty Zarr store of the proper dimensions at the target location.
Once this framework has been established, the algorithm moves on to actually copying bytes from the source data into the Zarr store, via the store_chunks task. Internally, this step performs a lot of heavy lifting, insofar as it determines which specific byte ranges within which source files are required to build each output chunk. Because both the cached source bytes and target dataset reside on cloud object storage, which supports scalable parallel reads and writes, this computationally intensive step is designed to be executed in parallel; specifically, each store_chunks task can be executed in any order, without communication or synchronization needed between processes. Parallelization of this step is essential to Pangeo Forge's performance, given that ARCO datasets are often hundreds of gigabytes in size on the low end, and can easily reach multi-petabyte scale.
Following the mirroring of all source bytes into their corresponding Zarr chunks, the XarrayZarrRecipe algorithm concludes with a finalization step which consolidates the dataset's metadata into a single lightweight JSON object.
Duplicating bytes is a costly undertaking, both computationally, and because cloud storage on the order of terabytes is not inexpensive. This is a primary reason why sharing these ARCO datasets via publicly accessible cloud buckets is so imperative: a single copy per cloud region or multi-region zone can serve hundreds or thousands of scientists. A clear advantage of the HDFReferenceRecipe algorithm is that it does not require byte duplication, however it has certain limitations. This approach requires that the data provider's server support random access to source file subsets, a common but non-universal feature of HTTP and FTP servers. Because the bytes on the data provider's server are not duplicated, use of HDFReferenceRecipe precludes forms of data preprocessing which modify the data itself; only metadata preprocessing is supported. Finally, opening data stores created by this algorithm requires the Python package kerchunk, effectively preventing access from languages other than Python, as of writing. The interface specification for virtual Zarr stores is clearly defined in the kerchunk documentation, therefore we anticipate it may be implemented in other languages in the future (Durant et al., 2021). HDFReferenceRecipe presents a remarkably efficient pathway for certain use cases, however as with most efficiencies, it comes with inevitable tradeoffs.
Pangeo Forge's initial algorithms produce Zarr outputs because this format is well-suited to ARCO representation of the gridded multidimensional array data that our early scientific adopters use in their research. Disadvantages of Zarr include the fact that popular data science programming languages such as R do not yet have an interface for the format (Durbin et al., 2020). As our community grows, we anticipate future recipe implementations to include support for most common ARCO formats. These include TileDB Embedded, for multidimensional arrays; Cloud Optimized GeoTIFF (COG), which is widely used in the geospatial imagery community; Parquet, for optimized tabular data stores; and the recently announced Cloud Optimized Point Cloud (COPC) format for, among other uses, light detection and ranging (LiDAR) measurements (Holmes, 2021; Le Dem and Blue, 2021; Hobu, Inc., 2021; TileDB, Inc., 2021). As with our Zarr algorithms, which depend on Xarray as an I/O interface, our path to implementing these algorithms will build on the standard Python interfaces for each data structure; e.g., Rasterio for raster data and Pandas for tabular data (Gillies et al., 2013; Pandas Development Team, 2021).
3.2.3. Storage Abstractions
In the discussion of source file patterns, above, we referred to the fact that input data may be arbitrarily sourced from a variety of different server protocols. The backend file transfer interface which enables this flexibility is the Python package Filesystem Spec, which provides a uniform API for interfacing with a wide range of storage backends (Durant, 2021). This same package provides the engine behind our storage abstractions, a set of modular components which handle various permutations of file caching, reading, and writing. These classes need not be enumerated here; the interested reader can find details about them in the Pangeo Forge documentation. One aspect of these components worth highlighting, however, is that even though cloud object storage is the typical destination of datasets processed by Pangeo Forge, the platform is just as easily able to read from and write to a local POSIX file system or, for that matter, any Filesystem Spec-compatible storage location. Among other things, this capability allows recipe contributors to experiment with recipe algorithms by writing ARCO dataset subsets to local disk during the development process. For our typical cloud storage interfaces, the Filesystem Spec implementations we employ most frequently are s3fs (for Amazon Web Services S3), gcsfs (for Google Cloud Storage), and adlfs (for Azure Datalake and Azure Blob Storage).
3.2.4. Execution Modes
Instantiating a recipe class does not by itself result in any data transformation actually occurring; it merely specifies the steps required to produce an ARCO dataset. In order to actually perform this workflow, the recipe must be executed. A central goal of the software design of pangeo-forge-recipes is to be as flexible as possible regarding the execution framework. A wide range of frameworks for parallel and/or distributed computing exist, and pangeo-forge-recipes seeks to be compatible with as many of these as possible. For example, high-performance computing (HPC) users may prefer to use traditional job-queue based execution, while cloud users may want to use Kubernetes (Brewer, 2015).
pangeo-forge-recipes does not directly implement any parallel computing. Rather, the library has the ability to compile recipes into several different formats used by common distributed computing frameworks. As of writing, we currently support four different flavors of compilation:
• Compilation to a single Python function: This is a reference implementation for serial execution.
• Compilation to Dask Delayed graph: Dask is a general purpose parallel computing framework widely used in the scientific Python world (Dask Development Team, 2016). By compiling recipes to Dask graphs, pangeo-forge-recipes users are able to leverage the variety of different schedulers Dask has implemented for a wide range of different computing platforms. These include dask-jobqueue for HPC systems using PBS, SLURM, SGE, etc. (Henderson, 1995; Gentzsch, 2001; Yoo et al., 2003); Dask Kubernetes for cloud; and Dask-Yarn for Hadoop (Shvachko et al., 2010). Dask's single machine schedulers enable recipes to be executed in parallel using threads or processes on a single large server.
• Compilation to Prefect Flow: Prefect is a suite of workflow automation tools encompassing both open source and software-as-a-service (SaaS) components: Prefect Core is an open source workflow engine for Python; a Prefect Flow is a set of interrelated individual tasks, structured in a graph; and Prefect Cloud is a SaaS platform which helps manage and monitor Flow execution (Prefect Technologies, Inc., 2021). Prefect provides a robust and observable way of running recipes and is our current default model for the Pangeo Forge cloud automation.
• Compilation to Apache Beam Pipeline: Apache Beam is an open source framework for defining parallel processing pipelines for batch and streaming data (Apache Software Foundation, 2016). Beam Pipelines are high-level dataflow graphs, composed of distributed datasets and globally optimized, lazily evaluated processing steps. By compiling to Beam Pipelines, pangeo-forge-recipes can be executed on major distributed computation systems including Apache Spark (Zaharia et al., 2016), Apache Fink (Apache Software Foundation, 2015), and Google Cloud Dataflow (Akidau et al., 2015), as well as through intermediaries such as Hadoop, Yarn, Mesos, and Kubernetes (Shvachko et al., 2010; Hindman et al., 2011; Brewer, 2015). Beam is a multi-language framework capable of executing multiple languages in a single Pipeline. This makes it possible to incorporate recipes into execution workflows outside of the Python ecosystem.
In addition to these execution frameworks, recipe steps can be manually run in sequential fashion in a Jupyter Notebook or other interactive environment (Ragan-Kelley et al., 2014). This facilitates user introspection and debugging.
3.3. Cloud Automation Platform
The nuclei of Pangeo Forge cloud automation are Bakeries, cloud compute clusters dedicated specifically to executing recipes. Bakeries provide a setting for contributors to run their recipes on large-scale, distributed infrastructure and deposit ARCO datasets into performant publicly-accessible cloud storage, all entirely free of cost for the user. By running their recipes in a Bakery, contributors are not only gaining access to free compute and storage for themselves, but are also making a considerable contribution back to the global Pangeo Forge community in the form of ARCO datasets which will be easily discoverable and reusable by anyone with access to a web browser.
Pangeo Forge follows the example of Conda Forge in managing its contribution process through the cloud-hosted version control platform GitHub. Recipe contributors who wish to run their recipes in a Bakery first submit their draft recipes via a Pull Request (PR) to the Pangeo Forge staged-recipes repository which, as the name implies, is a holding area for incoming recipes. Following an iterative review process, described in detail below, recipe PRs are approved by Pangeo Forge maintainers, at which point their contents are automatically transferred out of the staged-recipes repository and incorporated into a new, standalone repository known as a Feedstock. It is from this Feedstock repository that recipe execution is dispatched to the Bakery compute cluster. The details of and rationale behind this workflow are provided in the following subsections.
3.3.1. Contribution Workflow
Continuous integration (CI) is a software development practice whereby code contributions are reviewed automatically by a suite of specialized test software prior to being incorporated into a production codebase. CI improves code quality by catching errors or incompatibilities that may escape a human reviewer's attention. It also allows code contributions to a large project to scale non-linearly to maintainer effort. Equipped with a robust CI infrastructure, a single software package maintainer can review and incorporate large numbers of contributions with high confidence of their compatibility with the underlying codebase.
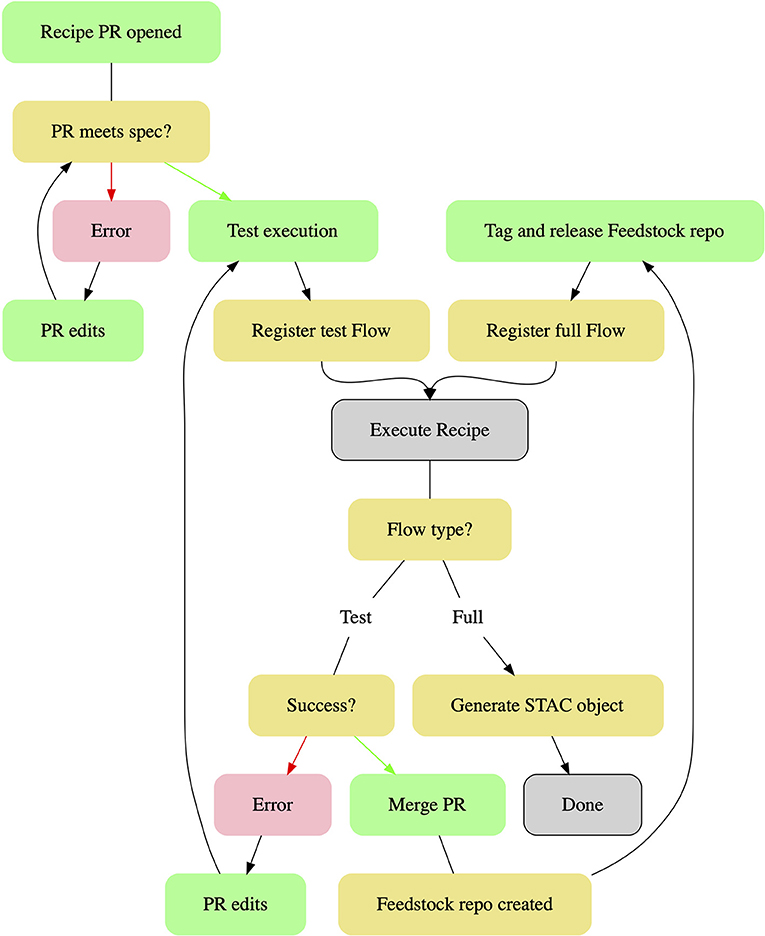
Pangeo Forge currently relies on GitHub's built-in CI infrastructure, GitHub Actions, for automated review of incoming recipe PRs (Figure 4). The first stage of this review process consists of checks that the submitted files, including the meta.yaml metadata and the recipe.py algorithm module, conform to the technical and stylistic specifications defined in the Pangeo Forge documentation. If errors are identified at this stage, the contributor is notified automatically and given a list of recommended changes, which must be incorporated prior to advancing to the next stage of evaluation.
Once the PR passes this first gate, a human project maintainer dispatches a command to run an automated execution test of the recipe. This test of a reduced subset of the recipe runs the same Prefect workflows on the same Bakery infrastructure which will be used in the full-scale data transformation. Creation of the reduced recipe is performed by a Pangeo Forge function which prunes the dataset to a specified subset of increments in the concatenation dimension. Any changes required to the recipe's functionality are identified here. For datasets expected to conform to Climate and Forecast (CF) Metadata Conventions, we plan to implement compliance checks at this stage using established validation tooling such as the Centre for Environmental Data Analysis (CEDA) CF Checker and the Integrated Ocean Observing System (IOOS) Compliance Checker (Adams et al., 2021; Eaton et al., 2021; Hatcher, 2021). Following an iterative process of corrections based on the results of the automated execution test (or a series of such tests, as necessary), the recipe PR is accepted by a human maintainer. At this point, a Feedstock repository is programmatically generated by incorporating the recipe PR files into a predefined repository template.
Creation of a Feedstock repository from the recipe PR triggers the full build of the ARCO dataset, after which the only remaining step in the contribution workflow is the generation of a catalog listing for the dataset, an automated process dispatched by GitHub Actions.
3.3.2. Feedstocks
Feedstocks are GitHub repositories which place user-contributed recipes adjacent to Pangeo Forge's cloud automation tools and grant access to Pangeo Forge credentials for authentication in a Bakery compute cluster. The Feedstock repository approach mirrors the model successfully established in Conda Forge. Those familiar with software version control processes will know that, most often, merging a PR results in proposed code changes being incorporated into an existing repository's codebase. As in Conda Forge, merging a PR to staged-recipes takes on a slightly different meaning in Pangeo Forge. Rather than incorporating a recipe's code into staged-recipes, merging a recipe PR results in the creation of a new, dedicated GitHub repository for the recipe called a Feedstock.
We can think of this new Feedstock repository as the deployed or productionalized version of the recipe. The template from which GitHub Actions automatically generates this repository includes automation hooks which register the recipe's ARCO dataset build with the specified Bakery infrastructure. All of these steps are orchestrated automatically by GitHub Actions and abstracted from the recipe code itself. As emphasized throughout this paper, this separation of concerns is intended to provide a pathway for scientific domain experts to participate in ARCO data curation without the requirement that they understand the highly-specialized domain of cloud infrastructure automation.
As public GitHub repositories, Feedstocks serve as invaluable touchstones for ARCO dataset provenance tracking. Most users will discover datasets through a catalog (more on cataloging in section 3.4). Alongside other metadata, the catalog entry for each dataset will contain a link to the Feedstock repository used to create it. This link connects the user to the precise recipe code used to produce each ARCO dataset. Among other benefits, transparent provenance allows data users to investigate whether apparent dataset errors or inconsistencies originate in the archival source data, or are artifacts of the ARCO production process. If the latter, the GitHub repository provides a natural place for collaboration on a solution to the problem. Each time a Feedstock repository is tagged with a new version number, the recipe it contains is re-built to reflect any changes made since the prior version.
Pangeo Forge implements a two-element semantic versioning scheme for Feedstocks (and, by extension, the ARCO datasets they produce). Each Feedstock is assigned a version conforming to the format MAJOR.MINOR and beginning at 1.0. Increments to the minor version are made for changes which are likely to be backwards-compatible with user code that relies on an earlier version of the data. Such updates include metadata corrections, adding new variables, or extending the temporal range of existing variables. Major version increments are triggered for non-backwards-compatible edits such as changing existing variable names or revising preprocessing functions such that they alter existing variable arrays. Pangeo Forge will maintain prebuilt copies of all major versions of a given dataset, but in the interest of storage efficiency will only retain the latest minor version for each of these major versioned datasets. (For example, if prebuilt copies of 1.0 and 2.0 exist in storage when 2.1 is released, 1.0 will be retained but 2.0 will be overwritten by 2.1.) In cases where a user may have a need for a specific minor version of a dataset that has already been superseded in storage by a new minor version release, the corresponding Feedstock can be used to rebuild any version of the ARCO data store on an as-needed basis.
3.3.3. Bakeries: On-Demand Cloud Clusters Paired With Cloud Storage Targets
As in Conda Forge, the majority of Pangeo Forge users will not execute recipes themselves, but rather interact with recipe outputs which are pre-built by shared cloud infrastructure. As such, execution typically only occurs once per recipe (or, in the case of updated recipe versions, once per recipe version). This one-time execution builds the ARCO dataset to a publicly-accessible cloud storage bucket. Arbitrary numbers of data users can then access the pre-built dataset directly from this single shared copy. This approach has many advantages for our use case, including:
• Shared compute is provisioned and optimized by cloud infrastructure experts within our community to excel at the specific workloads associated with ARCO dataset production.
• As a shared resource, Pangeo Forge cloud compute can be scaled to be larger and more powerful than most community users are likely to be able to provide themselves.
• Storage and compute costs (financial, and in terms of environmental footprint) are not duplicated unnecessarily.
Costs for these shared resources are currently covered through a combination of free credits provided by technology service providers and grants awarded to Pangeo Forge.
Bakeries, instances of Pangeo Forge's shared cloud infrastructure, can be created on Amazon Web Services, Microsoft Azure, and Google Cloud Platform cloud infrastructure. In keeping with the aforementioned Right to Replicate, an open source template repository, tracing a clear pathway for reproducing our entire technology stack, is published on GitHub for each supported deployment type (2i2c.org, 2021). In practice, the cost and complexity of these deployments likely means they will be undertaken by organizations rather than individuals. Over time, we anticipate the benefits of participating in Pangeo Forge will motivate a wide range of both non-profit and commercial partners to establish Bakeries for community use. The greater the number and scale of Bakeries in operation, the greater the capacity of Pangeo Forge to democratize the means of ARCO data production.
When a community member submits a Pangeo Forge recipe, they use the meta.yaml file included as part of each recipe submission to specify the Bakery on which to execute it, and the target storage location within that Bakery in which to deposit the resulting dataset. Each Bakery will manage their own compliment of storage buckets. Available Bakeries and their specifications, including storage bucket protocols and locations, are recorded in a public database for reference. Selection of one Bakery over another may be based on factors including the geographic location of the associated storage bucket(s), given that physical proximity of compute resources to data impacts performance for big data analytics.
3.4. Cataloging and Loading
The SpatioTemporal Asset Catalog (STAC) is a human and machine readable cataloging standard gaining rapid and broad traction in the geospatial and earth observation (EO) communities (Alemohammad, 2019; Emanuele, 2020; Holmes et al., 2021). The value of STAC is enhanced by its tooling ecosystem, which includes interfaces for many programming languages and a community-supported web frontend (Emanuele et al., 2021; Fitzsimmons et al., 2021). STAC was not originally conceived as a cataloging solution for the Earth-system model (ESM) data which will constitute a majority of Pangeo Forge's ARCO data holdings, however extensions such as the Datacube Extension bring descriptive cataloging of ESM data with STAC within reach (Mohr et al., 2021). Despite the imperfect fit of ESM data into STAC, the momentum behind this specification and its associated ecosystem recommends it as the best option for implementation of our user-facing catalog.
Following the completion of each ARCO production build, GitHub Actions automatically generates a STAC listing for the resulting dataset and adds it to the Pangeo Forge root catalog. Information which can be retrieved from the dataset itself (including dimensions, shape, coordinates, and variable names) is used to populate the catalog listing whenever possible. Fields likely not present within the dataset, such as a long description and license type, are populated with values from the meta.yaml file which contributors include as part of each recipe.
STAC provides not only a browsing interface, but also defines a streamlined pathway for loading datasets. Catalog-mediated loading simplifies the user experience as compared to the added complexity of loading directly from a cloud storage Uniform Resource Identifier (URI). Pangeo Forge currently provides documentation for loading datasets with Python into Jupyter Notebooks, given that our early adopters are likely to be Python users (Perkel, 2018). One distinct advantage of STAC's JSON-based specification over other language-specific cataloging options is its current (or in some cases, planned) interoperability with a wide variety of programming languages. We look forward to documenting catalog access from JavaScript, R, Julia, and many other contemporary languages as our user community grows.
Discoverability is the ease with which someone without prior knowledge of a particular dataset can find out about its existence, locate the data, and make use of it. As the project grows, we aspire to enhance data discoverability by offering a range of search modalities for the Pangeo Forge ARCO dataset catalog, enabling users to explore available datasets by spatial, temporal, and variable extents.
4. Examples
In the course of development and validation, we employed Pangeo Forge to transform a selection of archival NetCDF datasets, collectively totalling more than 2.5 terabytes in size, into the cloud-optimized Zarr format. The resulting ARCO datasets were stored on the Open Storage Network (OSN), an NSF-funded instance of Amazon Web Services S3 storage infrastructure, and have already been featured in multiple presentations and/or played a central role in ongoing research initiatives. We offer a brief summary of these example results below followed by some general reflections, drawn from these experiences, on the performance of the platform to date.
4.1. SWOT Ocean Model Intercomparison
The upcoming Surface Water and Ocean Topography (SWOT) satellite mission will measure sea-surface height at high resolution with synthetic aperture radar (Morrow et al., 2019). In coordination with this mission, an international consortium of oceanographers are currently undertaking modeling and in-situ field campaigns for purposes of comparison to the forthcoming SWOT satellite measurements (Li, 2019). As part of these efforts, we have transformed portions of the outputs from the FESOM, GIGATL, HYCOM, eNATL60, and ORCA36 ocean models into ARCO datasets with Pangeo Forge (Chassignet et al., 2007; Danilov et al., 2017; Brodeau et al., 2020; Castrillo, 2020; Gula, 2021). From a technical perspective, these transformations involved caching approximately a terabyte of ocean model data from FTP servers in France and Germany onto Google Cloud Storage in Iowa, USA via Pangeo Forge's internal file transfer utilities. This experience highlighted the persisting influence of geographic distance on network communication speeds and led to many improvements in how we manage file transfer internally within the platform. From the standpoint of data structure, the multigigabyte-scale array sizes contained within some of these model outputs encouraged the development of a specialized subsetting pathway within pangeo-forge-recipes for handling larger-than-memory input arrays.
4.2. NOAA Optimal Interpolation Sea Surface Temperature
NOAA's Optimal Interpolation Sea Surface Temperature (OISST) is a daily resolution data product combining in-situ field measurements with satellite temperature observations from the Advanced Very High Resolution Radiometer (AVHRR) (Huang et al., 2021). With Pangeo Forge, we created a single consolidated Zarr store from 14,372 NOAA OISST source files spanning a time range from 1981 to 2021. This Zarr store was subsequently used as part of investigations into the morphology of ocean temperature extremes (Scannell et al., 2021). In many ways, this flavor of recipe (concatenation of NetCDF timeseries archives into a consolidated ARCO store) is what the earliest versions of Pangeo Forge were designed to excel at. We therefore relied heavily on this recipe during early development as a useful test case for our cloud automation infrastructure.
4.3. CMEMS Sea Surface Altimetry
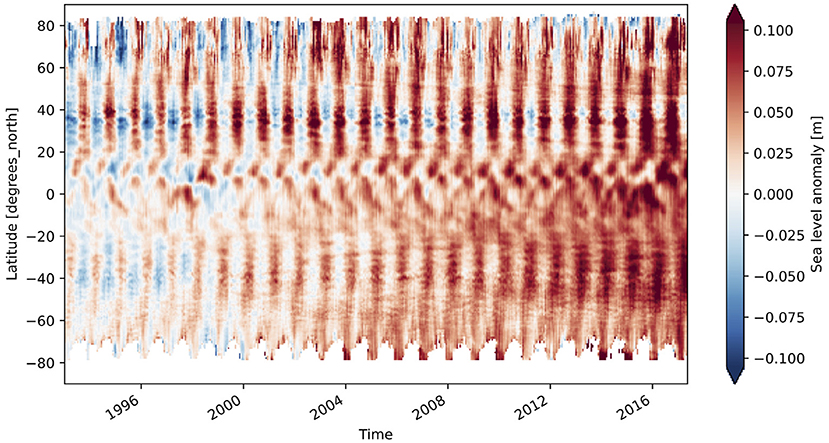
A 70 gigabyte ARCO dataset of gridded sea surface altimetry measurements was assembled by Pangeo Forge from nearly 9,000 files sourced from the Copernicus Marine Service (Copernicus Marine Environment Monitoring Service, 2021). For researchers wishing to study trends in sea level, downloading so many files is a laborious barrier to science. With the Pangeo Forge ARCO dataset, a reduction over the entire dataset to visualize the global patterns of sea-level rise can be accomplished in less than a minute and with just a few lines of code (shown in Listing 3). This calculation was performed as part of live demonstrations of Pangeo Forge presented at recent ESIP and Research Running on Cloud Compute and Emerging Technologies (RRoCCET) conferences (Barciauskas et al., 2021; Stern, 2021).
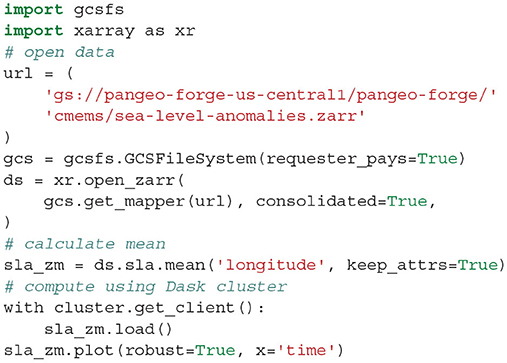
Listing 3. Code used to generate Figure 5 from the Pangeo Forge ARC sea-level data.
4.4. CESM POP 1-Degree
Processing this low-resolution output of the Community Earth System Model (CESM) became an unexpected but welcome opportunity to examine how Pangeo Forge handles user credentials for accessing source files and resulted directly in the addition of query string authentication features to pangeo-forge-recipes. Regarding the data transformation itself, the source files for this recipe represented yet another example of containing larger-than-memory variable arrays (National Center for Atmospheric Research, 2021). The development team's swift and successful adaptation of Pangeo Forge to accommodate this use case is a testament to the extensibility of the platform's base abstractions.
4.5. SODA 3.4.2 ICE
The Simple Ocean Data Assimilation (SODA) model aims to reconstruct twentieth century ocean physics (Carton et al., 2018). We transformed a subset of this model's output consisting of roughly 2,100 source files into a consolidated ARCO data store to aid a colleague's ongoing research.
4.6. Challenges, Performance, and Costs
We have had no difficulty converting any of the NetCDF files from our use cases into Zarr, thanks to Xarray's sophisticated metadata and encoding management. Xarray faithfully replicates all variables, metadata, and datatypes present in the archival NetCDF files into their Zarr analogs such that the resulting Zarr stores, when opened with Xarray, are identical to the dataset present in the original (aggregated) NetCDF files. The main known limitation of Xarray's Zarr interface is that it does not support hierarchically nested NetCDF groups, only flat groups; this particular limitation has not affected our above-listed use cases.
Pangeo Forge is generally I/O bound. The greatest challenge we have experienced is slow downloading from source data archives during the caching phase of recipe execution. If too many simultaneous requests are made to an HTTP or FTP source server, this will typically result in the per-file transfer throughput decreasing considerably. Therefore, caching source files for a given recipe is not highly parallelizable. As noted in section 4.1, transcontinental data transfer can be slow process, even for sequential requests. Once the source files are cached into the cloud, however, platform performance scales out well, since all I/O is happening against cloud object storage.
We have not yet made a systematic assessment of cost and performance. Regarding minimum hardware requirements, Bakery workloads are typically distributed across large numbers of lightweight compute nodes. In a typical implementation, each node may be provisioned with roughly 4 gigabytes of RAM and one CPU core. The larger-than-memory archival arrays referenced in sections 4.1, 4.4 challenged this computational model and prompted the addition of subsetting routines to the platform that facilitate division of arbitrarily-sized input arrays along one or multiple dimensional axes. This allows our lightweight compute nodes to handle inputs in excess of their RAM allocation. As we move from the initial software development phase into productionalization of increasing numbers of Bakeries, we look forward to sharing more fine-grained assessments of the platform's performance and resource requirements.
5. Future Outlook
As of the time of writing this paper, all of the major components of Pangeo Forge (with the exception of the data catalog) have been released openly on GitHub, tested thoroughly, and integrated through end-to-end workflows in the cloud. Dozens of actual and potential users have interacted with the project via GitHub issues and bi-weekly meetings. However, the platform has not been officially “launched,” as in, advertised broadly to the public as open for business. We anticipate taking this step in early 2022. After that point, development will continue indefinitely into the future as we continue to refine and improve the service in response to user feedback.
The current development of Pangeo Forge is supported by a 3 year grant from the National Science Foundation (NSF) EarthCube program. Storage expenses are covered through our partnership with the Open Storage Network (OSN), which provides Pangeo Forge with 100 terabytes of cloud storage space, accessible over the S3 protocol for free (Public cloud storage buckets often implement a “requester-pays” model in which users are responsible for the cost of moving data; our OSN storage does not). All three major cloud providers offer programs for free hosting of public scientific datasets. We anticipate engaging in these programs as our storage needs grow. We have also begun to evaluate distributed, peer-to-peer storage systems such as the InterPlanetary FileSystem (IPFS) and Filecoin as an alternative storage option.
Pangeo Forge is not itself an archival repository but rather a platform for transforming data, sourced from archival repositories, into optimized formats. We therefore do not commit to preserving every recipe's materialized data in perpetuity. In nearly all cases, recipes source data from archival repositories with a long-term stewardship plan. It should therefore almost always be possible to regenerate a Pangeo Forge ARCO dataset by re-running the recipe contained in the versioned Feedstock repository from which it was originally built.
We hope that the platform we create during the course of our NSF award will gain traction that merits long-term financial support for the project. The level of support required will depend on the volume of community interest and participation. In any scenario, however, it is not feasible for Pangeo Forge core development team to personally maintain every Feedstock. Instead, via the crowdsourcing model, we aspire to leverage the expertise of a large community of contributors, each of whom will be responsible for keeping their recipes up to date. The core team will support these maintainers to the greatest degree possible, via direct mentorship as well as more scalable modes of support such as documentation and automated integration tests. Recipe contribution is not the only thing we envision crowdsourcing. Indeed, the platform itself is designed to be “franchisable”: any organization can run a Bakery. We envision Pangeo Forge not as a single system with one owner but rather as a federation. Participating organizations will bear the compute and storage costs of the datasets they care about supporting and recipes will be routed to an appropriate Bakery as part of the GitHub contribution workflow.
In the remainder of this final section, we conclude by imagining a future state, several years from now, in which Pangeo Forge has cultivated a broad community of recipe contributors from across disciplines, who help populate and maintain a multi-petabyte database of ARCO datasets in the cloud. How will this transform research and applications using environmental data? What follows is inherently speculative, and we look forward to revisiting these speculations in several years time to see how things turn out.
5.1. An Ecosystem for Open Science
Pangeo Forge and the ARCO data repositories it generates are most valuable as part of a broader ecosystem for open science in the cloud (Gentemann et al., 2021). In particular, Pangeo Forge ARCO data is designed to be used together with scalable, data-proximate computing. For interactive data analysis, Jupyter (including Jupyter Lab and Jupyter Hub) is emerging as a consensus open-source platform for the scientific community (Kluyver et al., 2016). Jupyter supports interactive computations in all major scientific computing languages, including Python, R, and Julia (We note especially that, although Pangeo Forge itself is written in Python, the data formats and catalogs it generates are all based on open standards, accessible from any major programming language). Jupyter in the cloud, combined with cloud-native parallel computing tools such as Dask (Rocklin, 2015) and Spark (Zaharia et al., 2016), creates a complete end-to-end solution for data-intensive research based purely on open-source software. By accelerating the production and sharing of ARCO data, we hope to stimulate further development and broad adoption of this new model for scientific research.
Beyond expert analysis, we also hope that the datasets produced by Pangeo Forge will enable a rich downstream ecosystem of tools to allow non-experts to interact with large, complex datasets without writing code. ARCO formats like Zarr are ideal for powering APIs, dashboards, and interactive websites, since they are based on open standards and can be read quickly from any programming language, including JavaScript, the language of the web. As an example, the sea-level data shown in Figure 5 could be used to create an interactive data visualization website for high-school students to study sea level change. Students wishing to go beyond the visual exploration could transition to an interactive Jupyter notebook and write their first lines of code, all pointing at the same underling data. Similarly, industry experts and policy makers could use such tools to examine climate impacts on their sector of interest. The direct provenance chain from the interactive tool, to the ARCO data copy, to the original upstream data provider would provide a fully transparent and trustworthy foundation for decision making.
5.2. Collaboration and Recognition Around Data Production
While nearly all scientists recognize the importance of data for research, scientific incentive systems do not value data production nearly as much as other types of scientific work, such as model development (Pierce et al., 2019). This was emphasized in a recent paper from Google Research, warning of the impact of data quality issues in the context of artificial intelligence research (Sambasivan et al., 2021). The undervaluing of “data work” is pervasive in the sciences, as evidenced by the existence of pejorative terms such as “data janitor.” Data work often occurs in the shadows of science, not talked about much in papers or recognized via honors and awards. One of our central hopes with Pangeo Forge is that the preparation of well curated, quality controlled datasets immediately accessible to high-performance computing will become an area of increased collaboration and visibility in environmental science research. By leveraging the interactivity inherent in GitHub discussions, we hope to see researchers from different institutions and countries coming together around building shared datasets of use to many different groups. By establishing a community storehouse of datasets themselves, as well as Feedstock repositories containing dataset provenances, we hope to offer citable artifacts of data production which, if reused and credited by the community, may serve to elevate the profile of this essential scientific work. Perhaps 1 day we will give an award for “most valuable recipe”!
Pangeo Forge does not currently implement a system for assigning unique persistent identifiers, such as Digital Object Identifiers (DOIs), to either Feedstocks or the datasets they produce. We certainly appreciate the tremendous benefit such identifiers provide, particularly for purposes of academic citation. As noted above, at this stage of our development we are not making a commitment to keeping datasets online in perpetuity, as would be required for a DOI. This reflection leads us to conclude that the Pangeo Forge Feedstock, a lightweight repository which will be permanently stored on GitHub, may in fact be the most appropriate object for DOI-assignment and citation. Feedstocks (and within them, recipes) are also the products which are most plainly expressive of contributors' technical and domain expertise. We welcome community feedback on how to best support contributors to receive the credit and recognition they deserve.
5.3. Asking More Ambitious Questions From Data
A recurring theme of the examples in section 4 is the relative simplicity of aligning thousands of source files into a single consolidated dataset with Pangeo Forge. The ARCO datasets which result from this process are not simply faster to work with than archival data, in many cases they enable an entirely new worldview. When working within the confines of traditional filesystems, it can be difficult for the scientific imagination to fly nimbly across the grand spatial and temporal scales permitted by ARCO workflows. By making entire worlds (observed or synthetic, past or future) accessible in an instant through shared ARCO data stores, we wholly expect that Pangeo Forge to not only accelerate existing science, but to also play a pivotal role in the reimagination of what's possible in ocean, weather, and climate science at scale.
5.4. Reproducibility in Action
Each Pangeo Forge recipe encodes data provenance starting from an archival source, all the way to the precise derived version used for a given research project. Tracking an unbroken provenance chain is particularly important in the context of ARCO data, which undergoes significant transformation prior to being used for analysis. The algorithms used to create ARCO datasets encode assumptions about what types of homogenization and/or simplification may serve the investigation for which the dataset is being produced. These judgement calls can easily be as impactful to the scientific outcome as the analysis itself. By tracking the ARCO production methodology through a recipe's Feedstock repository, Pangeo Forge affords visibility into the choices made at the data curation stage of research.
The oft-quoted eighty-twenty rule describes a typical ratio of time required for cleaning and preparing data vs. actually performing analysis. Depending on the type of preprocessing applied to a dataset, the time and technical knowledge required to reproduce previous derived datasets, let alone results, represents a major barrier to reproducibility in computational science. Duplication of data preparation is unnecessary and can be avoided if the dataset used for a given study, along with the recipe used to create it, are made publicly accessible.
5.5. Broadening Participation
Traditionally, working with big environmental datasets has required considerable infrastructure: big computers, hard drives, and IT staff to maintain them. This severely limits who can participate in research. One of the great transformative potentials of cloud-native science is the ability to put powerful infrastructure into the hands of anyone with an internet connection (Gentemann et al., 2021). In our recent experience, we have observed that it is easy enough to get started with cloud computing; the hard part is getting the right data into the cloud in the right format.
Pangeo Forge not only shifts the infrastructure burden of data production from local infrastructure to the cloud; it also lightens the cognitive burden for potential contributors by encouraging them to focus on the domain-specific details of the data, rather than the data engineering. As a recipe contributor to Pangeo Forge, anyone with a laptop can run their ARCO transformation algorithm at a scale previously only available to a small organizationally-affiliated group.
The true success of Pangeo Forge depends on creation of a space where a diverse community of recipe contributors can come together to curate the ARCO datasets which will define the next decade of cloud-native, big-data ocean, weather, and climate science. How we best nurture this community, and ensure they have the education, tools, and support they need to succeed, remains an open question, and an area where we seek feedback from the reader.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
CS drafted the manuscript with contributions from all other authors. All authors contributed to the design and implementation of Pangeo Forge. All authors read and approved the submitted version of the manuscript.
Funding
Pangeo Forge development is funded by the National Science Foundation (NSF) Award 2026932.
Conflict of Interest
AM was employed by company Google. SH was employed by company Development Seed.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Pangeo Forge benefits from the thoughtful contributions and feedback of a broad community of scientists and software engineers. We extend heartfelt thanks to all those who have contributed in any form including code, documentation, or via participation in community meetings and discussions.
References
2i2c.org (2021). The Customer Right to Replicate. Available online at: https://2i2c.org/right-to-replicate/ (accessed September 22, 2021).
Abernathey, R. P., Augspurger, T., Banihirwe, A., Blackmon-Luca, C. C., Crone, T. J., Gentemann, C. L., et al. (2021). Cloud-native repositories for big scientific data. Comput. Sci. Eng. 23, 26–35. doi: 10.1109/MCSE.2021.3059437
Adams, B., Campbell, L., Kell, D., Fernandes, F., Fratantonio, B., Foster, D., et al. (2021). IOOS Compliance Checker. Available online at: https://github.com/ioos/compliance-checker
Akidau, T., Bradshaw, R., Chambers, C., Chernyak, S., Fernández-Moctezuma, R. J., Lax, R., et al. (2015). The dataflow model: A practical approach to balancing correctness, latency, and cost in massive-scale, unbounded, out-of-order data processing. Proc. VLDB Endowment 8, 1792–1803. Available online at: https://research.google/pubs/pub43864/
Alemohammad, H. (2019). “Radiant mlhub: A repository for machine learning ready geospatial training data,” in AGU Fall Meeting Abstracts, (IN11A–05) (Washington, DC).
Anaconda Inc. (2021). Anaconda Software Distribution. Available online at: https://docs.anaconda.com/
Apache Software Foundation (2015). Apache Fink. Available online at: https://flink.apache.org/
Apache Software Foundation (2016). Apache Beam. Available online at: https://beam.apache.org/
Barciauskas, A., Shrestha, S., Casey, R., Signell, R., Friesz, A., Olson, S., et al. (2021). “The saga continues: cloud-optimized data formats,” in Earth Science Information Partners (ESIP) Summer Meeting 2021 (Severna Park, MD: ESIP).
Brewer, E. A. (2015). Kubernetes and the path to cloud native. in Proceedings of the Sixth ACM Symposium on Cloud Computing, SoCC '15, (Association for Computing Machinery) (New York, NY), 167.
Brodeau, L., Sommer, J. L., and Albert, A. (2020). Ocean-next/eNATL60: Material Describing the Set-up and the Assessment of NEMO-eNATL60 Simulations (Version v1). Zenodo. doi: 10.5281/zenodo.4032732
Carton, J. A., Chepurin, G. A., and Chen, L. (2018). Soda3: a new ocean climate reanalysis. J. Climate 31, 6967–6983. doi: 10.1175/JCLI-D-18-0149.1
Castrillo, M. (2020). “The nemo orca36 configuration and approaches to increase nemo4 efficiency,” in The 6th European Network for Earth System Modelling (ENES) Workshop on High Performance Computing for Climate and Weather (ENES). Available online at: https://www.esiwace.eu/events/6th-hpc-workshop/presentations/the-nemo-orca36-configuration-and-approaches-to-increase-nemo4-efficiency
Chassignet, E. P., Hurlburt, H. E., Smedstad, O. M., Halliwell, G. R., Hogan, P. J., Wallcraft, A. J., et al. (2007). The hycom (hybrid coordinate ocean model) data assimilative system. J. Marine Syst. 65, 60–83. doi: 10.1016/j.jmarsys.2005.09.016
Conda-Forge Community (2015). The Conda-Forge Project: Community-Based Software Distribution Built on the Conda Package Format and Ecosystem. Zenodo. doi: 10.5281/zenodo.4774217
Copernicus Marine Environment Monitoring Service (2021). Global Ocean Gridded l4 Sea Surface Heights and Derived Variables Reprocessed (1993-ongoing). Available online at: https://resources.marine.copernicus.eu/product-detail/SEALEVEL_GLO_PHY_L4_REP_OBSERVATIONS_008_047/INFORMATION
Cornillon, P., Adams, J., Blumenthal, M. B., Chassignet, E., Davis, E., Hankin, S., Kinter, J., et al. (2009). Nvods and the development of opendap. Oceanography 22, 116–127. doi: 10.5670/oceanog.2009.43
Danilov, S., Sidorenko, D., Wang, Q., and Jung, T. (2017). The finite-volume sea ice–ocean model (fesom2). Geosci. Model Develop. 10, 765–789. doi: 10.5194/gmd-10-765-2017
Dask Development Team (2016). Dask: Library for Dynamic Task Scheduling. Avaialable online at: https://dask.org
Durant, M. (2021). fsspec: Filesystem Interfaces for Python. Avaialable online at: https://filesystem-spec.readthedocs.io/
Durant, M., Sterzinger, L., Signell, R., Jelenak, A., Maddox, L., Bell, R., et al. (2021). kerchunk. Avaialable online at: https://github.com/fsspec/kerchunk
Durbin, C., Quinn, P., and Shum, D. (2020). Task 51-cloud-optimized format study. NTRS—NASA Technical Reports Server.
Dwyer, J. L., Roy, D. P., Sauer, B., Jenkerson, C. B., Zhang, H. K., and Lymburner, L. (2018). Analysis ready data: Enabling analysis of the landsat archive. Remote Sens. 10, 1363. doi: 10.3390/rs10091363
Eaton, B., Gregory, J., Drach, B., Taylor, K., Hankin, S., Blower, J., et al. (2021). NetCDF Climate and Forecast (CF) Metadata Conventions. Available online at: https://cfconventions.org/
Emanuele, R. (2020). “Using spatiotemporal asset catalogs (stac) to modularize end-to-end machine learning workflows for remote sensing data,” in AGU Fall Meeting Abstracts, (IN007–01) (Washington, DC).
Emanuele, R., Duckworth, J., Engmark, V., Kassel, S., Schwehr, K., Olaya, V., et al. (2021). PySTAC: A library for working with SpatioTemporal Asset Catalog in Python 3. Available online at: https://github.com/stac-utils/pystac
Fitzsimmons, S., Mohr, M., Emanuele, R., Blackmon-Luca, C., et al. (2021). STAC Browser: A Vue-Based STAC Browser for Static Catalogs and APIs. Available online at: https://github.com/radiantearth/stac-browser
Gentemann, C. L., Holdgraf, C., Abernathey, R., Crichton, D., Colliander, J., Kearns, E. J., et al. (2021). Science storms the cloud. AGU Adv., 2:e2020AV000354. doi: 10.1029/2020AV000354
Gentzsch, W. (2001). Sun grid engine: towards creating a compute power grid. in Proceedings First IEEE/ACM International Symposium on Cluster Computing and the Grid, 35–36. doi: 10.1109/CCGRID.2001.923173
Gillies, S. (2013). Rasterio: Geospatial Raster I/O for Python Programmers. Mapbox. Available online at: https://github.com/rasterio/rasterio
Gorelick, N., Hancher, M., Dixon, M., Ilyushchenko, S., Thau, D., and Moore, R. (2017). Google earth engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 202, 18–27. doi: 10.1016/j.rse.2017.06.031
Gula, J. (2021). Mesharou/GIGATL: Description of the GIGATL Simulations (v1.1). Zenodo. doi: 10.5281/zenodo.4948523
Hankin, S. C., Blower, J. D., Carval, T., Casey, K. S., Donlon, C., Lauret, O., et al. (2010). Netcdf-cf-opendap: Standards for ocean data interoperability and object lessons for community data standards processes. in Oceanobs 2009, Venice Convention Centre, 21-25 septembre 2009, Venise.
Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers, R., Virtanen, P., Cournapeau, D., et al. (2020). Array programming with NumPy. Nature 585, 357–362. doi: 10.1038/s41586-020-2649-2
Hatcher, R. (2021). cf-checker. Available online at: https://github.com/cedadev/cf-checker
Henderson, R. L. (1995). “Job scheduling under the portable batch system,” in Job Scheduling Strategies for Parallel Processing, eds D. G. Feitelson and L. Rudolph (Berlin; Heidelberg: Springer), 279–294.
Hindman, B., Konwinski, A., Zaharia, M., Ghodsi, A., Joseph, A. D., Katz, R. H., et al. (2011). Mesos: A platform for fine-grained resource sharing in the data center. in NSDI, vol. 11. 22–22. Available online at: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=+Mesos%3A+A+platform+for+fine-grained+resource+sharing+in+the+data+center.&btnG=
Hobu, Inc. (2021). Cloud Optimized Point CLoud (COPC). Available online at: https://copc.io/
Holmes, C. (2018). Analysis Ready Data Defined. Available online at: https://medium.com/planet-stories/analysis-ready-data-defined-5694f6f48815. (accessed September 09, 2021).
Holmes, C. (2021). Cloud Optimized GeoTIFF Specification. Available online at: https://github.com/cogeotiff/cog-spec
Holmes, C., Mohr, M., Hanson, M., Banting, J., Smith, M., Mathot, E., et al. (2021). SpatioTemporal Asset Catalog Specification-Making Geospatial Assets Openly Searchable and Crawlable. Available online at: https://github.com/radiantearth/stac-spec
Hoyer, S., and Hamman, J. (2017). xarray: N-D labeled arrays and datasets in Python. J. Open Res. Softw. 5, 10. doi: 10.5334/jors.148
Hua, H., Barciauskas, A., Chang, G., and Lynnes, C. (2020). “In042-lessons learned on supporting analysis ready data (ard) with analytics optimized data stores/services (aods) in collaborative analysis platforms posters,” in American Geophysical Union (AGU) Fall Meeting 2020 (Washington, DC: AGU).
Huang, B., Liu, C., Banzon, V., Freeman, E., Graham, G., Hankins, B., et al. (2021). Improvements of the daily optimum interpolation sea surface temperature (doisst) version 2.1. J. Climate 34, 2923–2939. doi: 10.1175/JCLI-D-20-0166.1
Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B., Bussonnier, M., Frederic, J., Kelley, K., Hamrick, J., Grout, J., Corlay, S., Ivanov, P., et al. (2016). “Jupyter notebooks a publishing format for reproducible computational workflows,” in Positioning and Power in Academic Publishing: Players, Agents and Agendas, eds F. Loizides and B. Scmidt (Amsterdam: IOS Press), 87–90. Available online at: https://www.iospress.com/catalog/books/positioning-and-power-in-academic-publishing-players-agents-and-agendas
Le Dem, J., and Blue, R. (2021). Apache Parquet. Available online at: https://github.com/apache/parquet-format
Li, J. (2019). SWOT Adopt-A-Crossover Consortium has been endorsed by CLIVAR. Available online at: https://www.clivar.org/news/swot-%E2%80%98adopt-crossover%E2%80%99-consortium-has-been-endorsed-clivar (accessed September 09, 2021).
Microsoft (2021). The Planetary Computer. Available online at: https://planetarycomputer.microsoft.com/
Miles, A., Bussonnier, M., Moore, J., Fulton, A., Bourbeau, J., Onalan, T., et al. (2021). zarr-developers/zarr-python: v2.10.3. Zenodo. doi: 10.5281/zenodo.5712786
Mohr, M., Hanson, M., Augspurger, T., Emanuele, R., Holmes, C., Scott, R., et al. (2021). Datacube Extension Specification. Available online at: https://github.com/stac-extensions/datacube
Morrow, R., Fu, L.-L., Ardhuin, F., Benkiran, M., Chapron, B., Cosme, E., et al. (2019). Global observations of fine-scale ocean surface topography with the surface water and ocean topography (swot) mission. Front. Marine Sci., 6:232. doi: 10.3389/fmars.2019.00232
National Center for Atmospheric Research. (2021). One degree, standard resolution CESM simulation from the Accelerated Scientific Discovery Phase of Yellowstone. NCAR Climate Data Gateway. Available Online at: https://www.earthsystemgrid.org/dataset/ucar.cgd.asd.cs.v5_rel04_BC5_ne30_g16.ocn.proc.daily_ave.html
Pangeo Forge Community (2021). Pangeo Forge. Available online at: https://pangeo-forge.readthedocs.io/
Perkel, J. M. (2018). Why jupyter is data scientists' computational notebook of choice. Nature 563, 145–147. doi: 10.1038/d41586-018-07196-1
Pierce, H. H., Dev, A., Statham, E., and Bierer, B. E. (2019). Credit data generators for data reuse. Nature 570, 30–32. doi: 10.1038/d41586-019-01715-4
Prefect Technologies Inc. (2021). Prefect. Available online at: https://docs.prefect.io/
Quinn, P., Abernathey, R., Signell, R., Neufeld, D., Privette, A., Killick, P., et al. (2020). “Cloud-optimized data,” in Earth Science Information Partners (ESIP) Summer Meeting 2020. ESIP.
Ragan-Kelley, M., Perez, F., Granger, B., Kluyver, T., Ivanov, P., Frederic, J., et al. (2014). “The jupyter/ipython architecture: a unified view of computational research, from interactive exploration to communication and publication,” in AGU Fall Meeting Abstracts, vol. 2014, H44D–07.
Ramamurthy, M. (2017). “Geoscience cyberinfrastructure in the cloud: data-proximate computing to address big data and open science challenges,” in 2017 IEEE 13th International Conference on e-Science (e-Science), 444–445.
Rew, R., Hartnett, E., Caron, J., et al. (2006). “Netcdf-4: software implementing an enhanced data model for the geosciences,” in 22nd International Conference on Interactive Information Processing Systems for Meteorology, Oceanography, and Hydrology, vol. 6.
Rocklin, M. (2015). “Dask: Parallel computation with blocked algorithms and task scheduling,” in Proceedings of the 14th python in science conference, vol. 130, 136. Citeseer.
Sambasivan, N., Kapania, S., Highfill, H., Akrong, D., Paritosh, P. K., and Aroyo, L. M. (2021). “Everyone wants to do the model work, not the data work”: Data cascades in high-stakes ai.
Scannell, H., Abernathey, R., Busecke, J., Gagne, D. J., Thompson, L., and Whitt, D. (2021). Ocetrac: morphological image processing for monitoring ocean temperature extremes. in Scientific Computing with Python (SciPy) 2021. SciPy.
Shvachko, K., Kuang, H., Radia, S., and Chansler, R. (2010). “The hadoop distributed file system,” in 2010 IEEE 26th Symposium on Mass Storage Systems and Technologies (MSST), 1–10.
Stall, S., Yarmey, L., Cutcher-Gershenfeld, J., Hanson, B., Lehnert, K., Nosek, B., et al. (2019). Make scientific data fair. Nature 570, 27–29. doi: 10.1038/d41586-019-01720-7
Stern, C. (2021). “Analysis ready data in the cloud,” in Research Running on Cloud Compute and Emerging Technologies (RRoCCET) 2021. RRoCCET. Available at Available online at: https://na.eventscloud.com/file_uploads/25629138ed86f9d6e6b4d8b8189e3b87_ConferenceProceedings.v2.pdf (accessed September 09, 2021).
TileDB, Inc. (2021). TileDB. Available online at: https://docs.tiledb.com/
Wagemann, J. (2020). ERA5 Reanalysis Data Available in Earth Engine. Available online at: https://www.ecmwf.int/en/newsletter/162/news/era5-reanalysis-data-available-earth-engine (accessed September 09, 2021).
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The fair guiding principles for scientific data management and stewardship. Sci. Data 3:160018. doi: 10.1038/sdata.2016.18
Yoo, A. B., Jette, M. A., and Grondona, M. (2003). “Slurm: Simple linux utility for resource management,” in Job Scheduling Strategies for Parallel Processing, eds D. Feitelson, L. Rudolph, and U. Schwiegelshohn (Berlin; Heidelberg: Springer), 44–60.
Keywords: data, community, cloud, ARCO, NetCDF, Zarr, Python
Citation: Stern C, Abernathey R, Hamman J, Wegener R, Lepore C, Harkins S and Merose A (2022) Pangeo Forge: Crowdsourcing Analysis-Ready, Cloud Optimized Data Production. Front. Clim. 3:782909. doi: 10.3389/fclim.2021.782909
Received: 25 September 2021; Accepted: 30 November 2021;
Published: 10 February 2022.
Edited by:
Michael C. Kruk, National Oceanic and Atmospheric Administration (NOAA), United StatesReviewed by:
Mark Capece, General Dynamics Information Technology, Inc., United StatesMIcah James Wengren, National Oceanic and Atmospheric Administration (NOAA), United States
Copyright © 2022 Stern, Abernathey, Hamman, Wegener, Lepore, Harkins and Merose. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryan Abernathey, rpa@ldeo.columbia.edu
 Charles Stern
Charles Stern Ryan Abernathey
Ryan Abernathey Joseph Hamman
Joseph Hamman Rachel Wegener
Rachel Wegener Chiara Lepore1
Chiara Lepore1